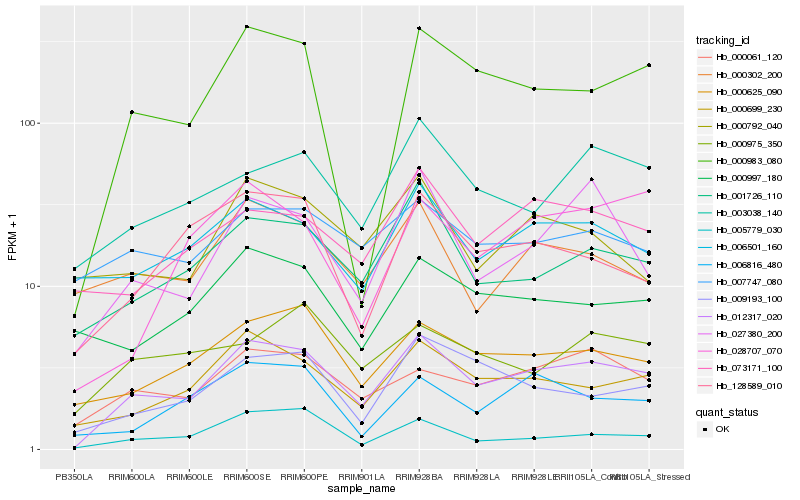
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012317_020 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_000699_230 |
0.1348007733 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 3 |
Hb_000983_080 |
0.1417616037 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 4 |
Hb_001726_110 |
0.1440635397 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 5 |
Hb_000061_120 |
0.1441151963 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 6 |
Hb_000625_090 |
0.1460601983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003038_140 |
0.1566103807 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 8 |
Hb_027380_200 |
0.1572561993 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_009193_100 |
0.1573465356 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 10 |
Hb_006501_160 |
0.1615457337 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 11 |
Hb_073171_100 |
0.161804295 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 12 |
Hb_128589_010 |
0.1653155661 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_005779_030 |
0.1678874806 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 14 |
Hb_000997_180 |
0.1721055757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007747_080 |
0.1729327982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006816_480 |
0.1731006587 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000302_200 |
0.173116603 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 18 |
Hb_028707_070 |
0.174091084 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000975_350 |
0.1755496744 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 20 |
Hb_000792_040 |
0.1777839094 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |