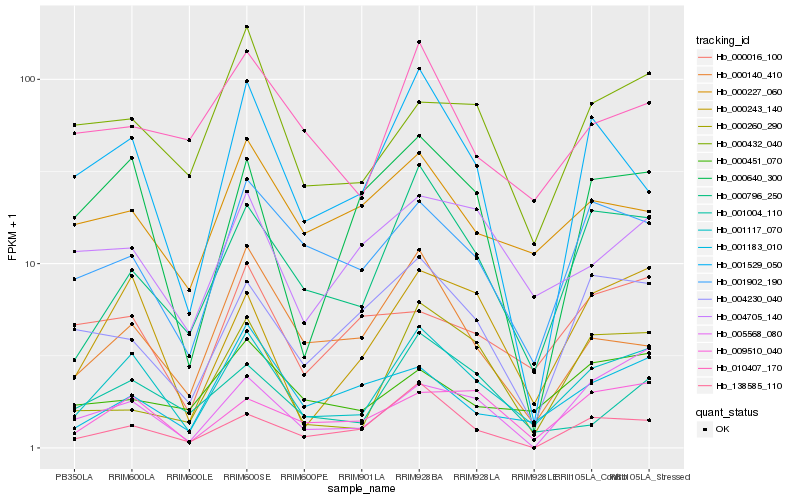
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_070 |
0.0 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 2 |
Hb_000243_140 |
0.1451739561 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 3 |
Hb_000796_250 |
0.1468501757 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 4 |
Hb_009510_040 |
0.1693798947 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001902_190 |
0.1814449002 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 6 |
Hb_000640_300 |
0.1873116903 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 7 |
Hb_000432_040 |
0.1920038543 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 8 |
Hb_001529_050 |
0.192243288 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 9 |
Hb_138585_110 |
0.19285834 |
- |
- |
- |
| 10 |
Hb_001004_110 |
0.1929658588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001183_010 |
0.1978093057 |
- |
- |
- |
| 12 |
Hb_004230_040 |
0.198563069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005568_080 |
0.198607575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004705_140 |
0.1987573503 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 15 |
Hb_010407_170 |
0.1987733354 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 16 |
Hb_000140_410 |
0.1990108805 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000227_060 |
0.2023004864 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 18 |
Hb_000451_070 |
0.203515477 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_000260_290 |
0.2044732658 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000016_100 |
0.2053940669 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |