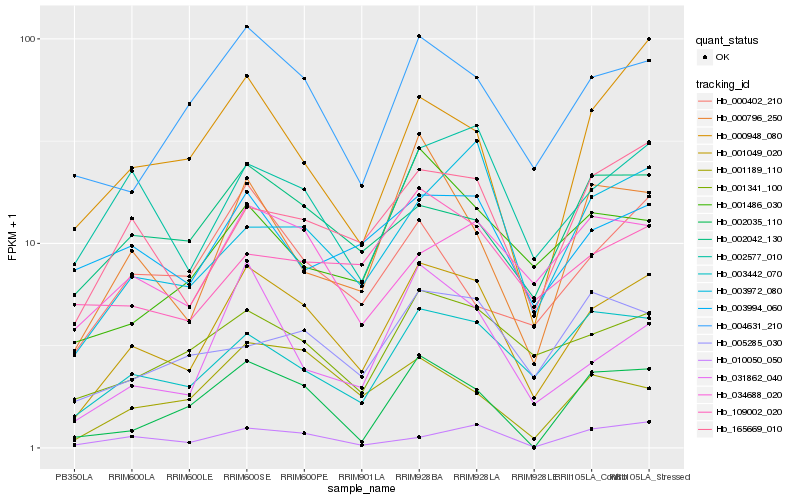
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001049_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004631_210 |
0.1393385912 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 3 |
Hb_001341_100 |
0.1447361015 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 4 |
Hb_003442_070 |
0.1459317726 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 5 |
Hb_165669_010 |
0.1477707787 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002577_010 |
0.1478017957 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 7 |
Hb_031862_040 |
0.1570174588 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 8 |
Hb_000796_250 |
0.1585511074 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 9 |
Hb_003972_080 |
0.1659675825 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 10 |
Hb_002042_130 |
0.1685919396 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 11 |
Hb_034688_020 |
0.1697493585 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 12 |
Hb_001486_030 |
0.1705612625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000948_080 |
0.1716027227 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 14 |
Hb_003994_060 |
0.1742393481 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 15 |
Hb_010050_050 |
0.1761138311 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_002035_110 |
0.1765249423 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 17 |
Hb_005285_030 |
0.1788453887 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 18 |
Hb_109002_020 |
0.1800945584 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 19 |
Hb_000402_210 |
0.1806229115 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 20 |
Hb_001189_110 |
0.1809363279 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |