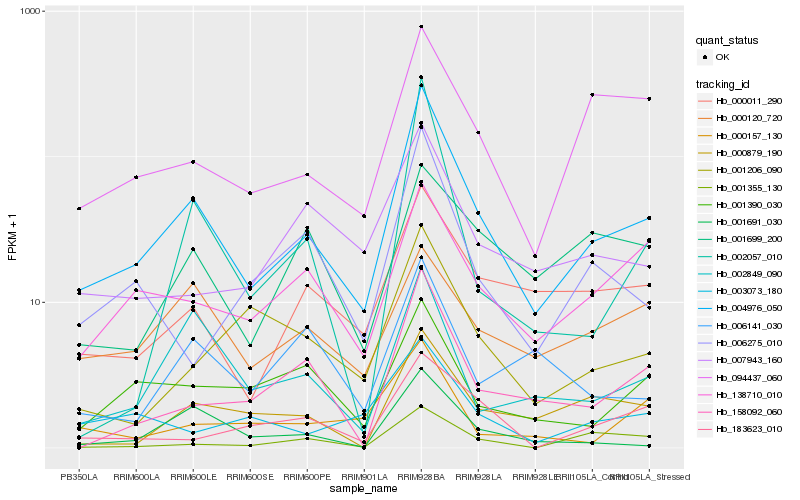
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004976_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000011_290 |
0.1998820082 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 3 |
Hb_138710_010 |
0.2007284629 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 4 |
Hb_094437_060 |
0.2042373238 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 5 |
Hb_002849_090 |
0.2074188097 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 6 |
Hb_000879_190 |
0.2140540559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_158092_060 |
0.2146720286 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_001691_030 |
0.21875361 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 9 |
Hb_001390_030 |
0.2203258418 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 10 |
Hb_007943_160 |
0.2210949328 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 11 |
Hb_006275_010 |
0.2218172939 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 12 |
Hb_001206_090 |
0.2267608491 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 13 |
Hb_003073_180 |
0.2315925653 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_002057_010 |
0.2320506707 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_001355_130 |
0.2328630223 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 16 |
Hb_000157_130 |
0.2353674874 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 17 |
Hb_000120_720 |
0.2355989562 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 18 |
Hb_183623_010 |
0.2361244922 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 19 |
Hb_006141_030 |
0.2388379117 |
- |
- |
- |
| 20 |
Hb_001699_200 |
0.242974929 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |