| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
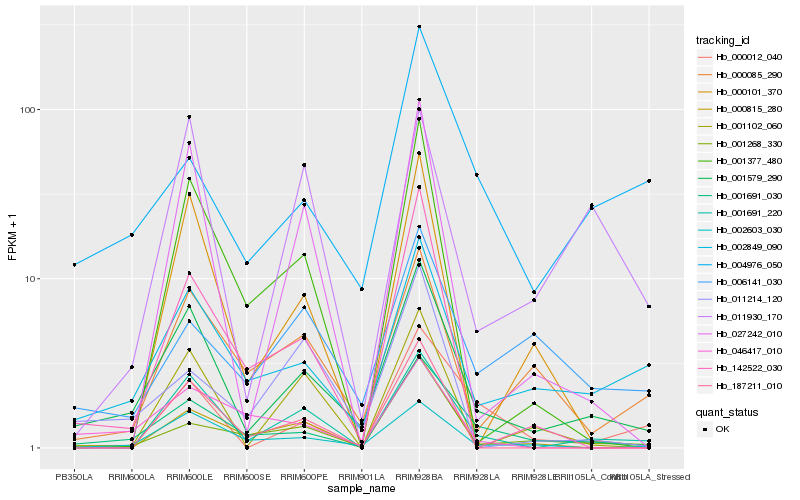
Hb_001579_290 |
0.0 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001691_030 |
0.1616450955 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 3 |
Hb_002849_090 |
0.1716669958 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 4 |
Hb_002603_030 |
0.1831641586 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_001691_220 |
0.1994670405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 6 |
Hb_000085_290 |
0.2254302667 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 7 |
Hb_001268_330 |
0.2296174914 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_027242_010 |
0.2398551443 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 9 |
Hb_142522_030 |
0.2421754042 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 10 |
Hb_000815_280 |
0.2426297408 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 11 |
Hb_004976_050 |
0.2441226475 |
- |
- |
- |
| 12 |
Hb_011930_170 |
0.2460304684 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 13 |
Hb_000101_370 |
0.2470737705 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_006141_030 |
0.2542080714 |
- |
- |
- |
| 15 |
Hb_001377_480 |
0.256432991 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 16 |
Hb_001102_060 |
0.259594519 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 17 |
Hb_046417_010 |
0.2596071751 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 18 |
Hb_187211_010 |
0.2600777033 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_011214_120 |
0.2611476966 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 20 |
Hb_000012_040 |
0.2612945946 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |