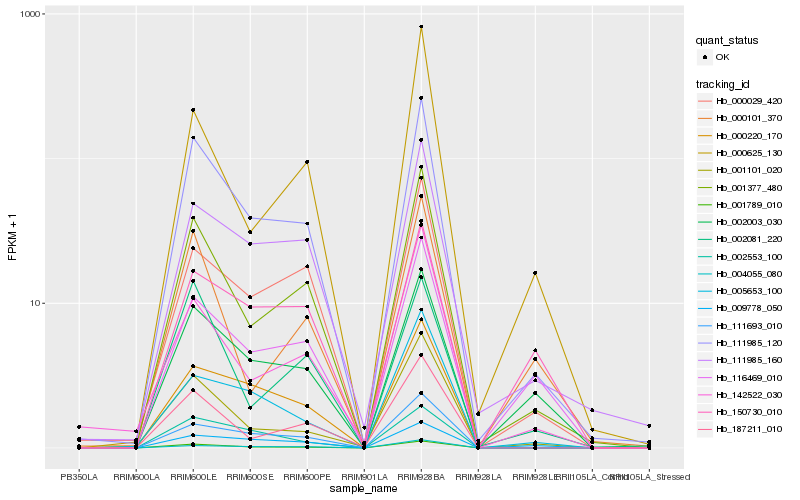
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111985_120 |
0.0 |
- |
- |
dihydroflavonol-4-reductase [Vitis rotundifolia] |
| 2 |
Hb_001789_010 |
0.0608804501 |
- |
- |
hypothetical protein POPTR_0015s13020g [Populus trichocarpa] |
| 3 |
Hb_001377_480 |
0.0834970937 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 4 |
Hb_111693_010 |
0.1100320691 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 5 |
Hb_000220_170 |
0.1163959622 |
- |
- |
- |
| 6 |
Hb_111985_160 |
0.1171833763 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 7 |
Hb_001101_020 |
0.1177234759 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 8 |
Hb_187211_010 |
0.1228210444 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 9 |
Hb_002003_030 |
0.1236278567 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 10 |
Hb_004055_080 |
0.1287753149 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_000029_420 |
0.1311802932 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 12 |
Hb_002553_100 |
0.1337667384 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 13 |
Hb_116469_010 |
0.142690085 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 14 |
Hb_005653_100 |
0.1457828085 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 15 |
Hb_142522_030 |
0.1546732215 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 16 |
Hb_002081_220 |
0.157367336 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 17 |
Hb_009778_050 |
0.1598579708 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 18 |
Hb_000101_370 |
0.1645187661 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000625_130 |
0.1661491443 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 20 |
Hb_150730_010 |
0.1681037235 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |