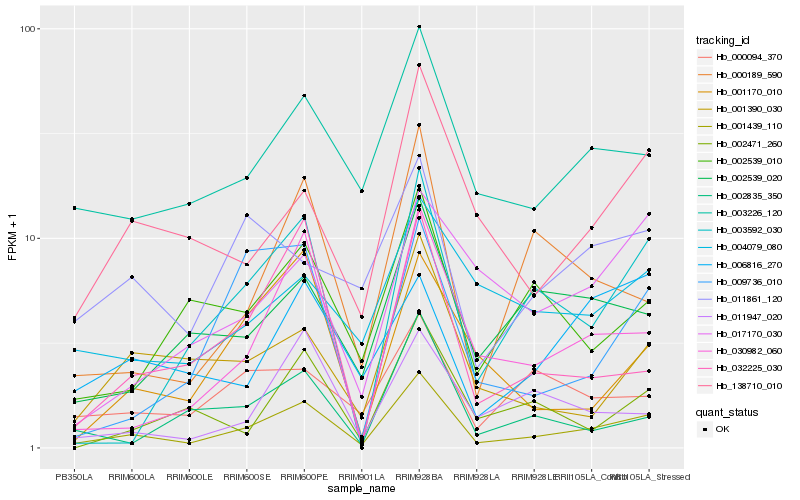
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001439_110 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 2 |
Hb_030982_060 |
0.1887931687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003226_120 |
0.1917373575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_138710_010 |
0.205136412 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 5 |
Hb_000189_590 |
0.2059722326 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 6 |
Hb_003592_030 |
0.20818704 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 7 |
Hb_001390_030 |
0.2134936489 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 8 |
Hb_002539_020 |
0.2135488767 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 9 |
Hb_032225_030 |
0.2135790037 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 10 |
Hb_002539_010 |
0.2204022128 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 11 |
Hb_011947_020 |
0.2212958628 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 12 |
Hb_009736_010 |
0.2234565404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002835_350 |
0.2259669751 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 14 |
Hb_000094_370 |
0.2269854553 |
- |
- |
- |
| 15 |
Hb_001170_010 |
0.2283275242 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_011861_120 |
0.2314904044 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 17 |
Hb_002471_260 |
0.2316516247 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 18 |
Hb_004079_080 |
0.2343820494 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 19 |
Hb_006816_270 |
0.2390359132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_017170_030 |
0.2397417872 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |