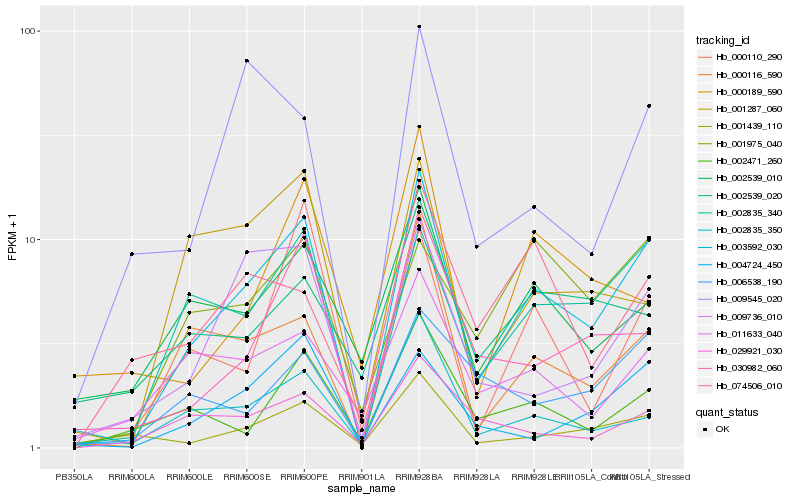
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003592_030 |
0.0 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 2 |
Hb_002835_340 |
0.1668885184 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 3 |
Hb_000116_590 |
0.1807726397 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 4 |
Hb_030982_060 |
0.1990961754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009736_010 |
0.2064206468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001439_110 |
0.20818704 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 7 |
Hb_004724_450 |
0.2130101784 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002471_260 |
0.2167202161 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 9 |
Hb_002539_010 |
0.2172888593 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 10 |
Hb_002835_350 |
0.219517574 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 11 |
Hb_011633_040 |
0.2198816152 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 12 |
Hb_029921_030 |
0.2225761111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001287_060 |
0.2260941986 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 14 |
Hb_006538_190 |
0.2273739432 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000110_290 |
0.2281420956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009545_020 |
0.2284295905 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 17 |
Hb_001975_040 |
0.2315335984 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 18 |
Hb_074506_010 |
0.2403318425 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 19 |
Hb_002539_020 |
0.2412092442 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 20 |
Hb_000189_590 |
0.2415756857 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |