| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
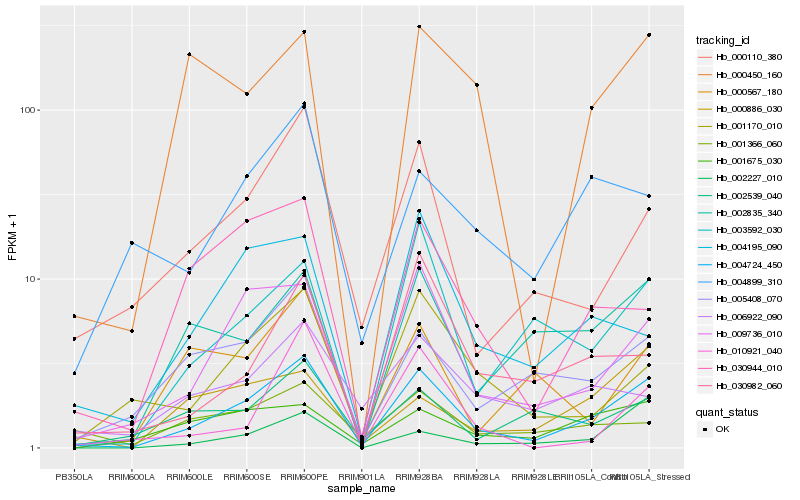
Hb_004724_450 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009736_010 |
0.1754796332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002835_340 |
0.2088283946 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 4 |
Hb_000110_380 |
0.2105291623 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 5 |
Hb_005408_070 |
0.2106647797 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 6 |
Hb_003592_030 |
0.2130101784 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 7 |
Hb_001170_010 |
0.2195069513 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000450_160 |
0.2297803324 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 9 |
Hb_001675_030 |
0.233997841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_030944_010 |
0.2346740645 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 11 |
Hb_002539_040 |
0.2360718688 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 12 |
Hb_030982_060 |
0.2364247435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004195_090 |
0.2375311382 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 14 |
Hb_004899_310 |
0.2420371615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000886_030 |
0.244906073 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 16 |
Hb_006922_090 |
0.2451311914 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 17 |
Hb_001366_060 |
0.2463411129 |
- |
- |
- |
| 18 |
Hb_000567_180 |
0.2471825268 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 19 |
Hb_002227_010 |
0.2475139853 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Populus euphratica] |
| 20 |
Hb_010921_040 |
0.2499620514 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |