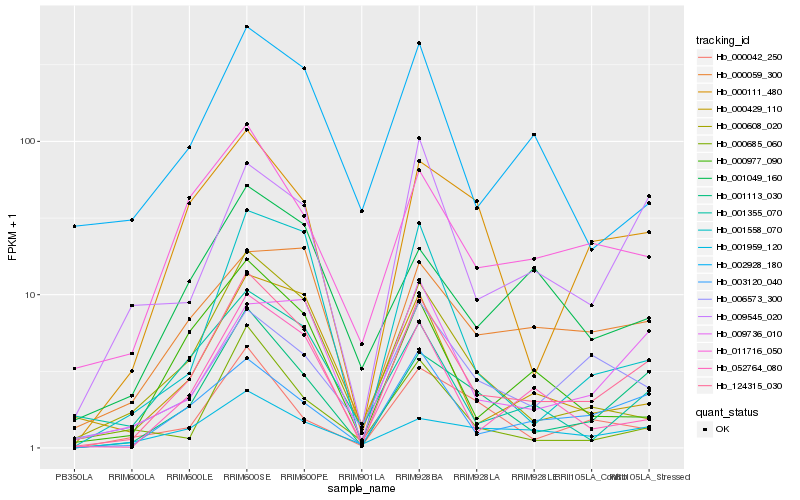
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124315_030 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 2 |
Hb_001113_030 |
0.1298226838 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 3 |
Hb_003120_040 |
0.1622139439 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 4 |
Hb_052764_080 |
0.1650340353 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000608_020 |
0.1779140256 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 6 |
Hb_011716_050 |
0.1793176766 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 7 |
Hb_009545_020 |
0.1885373619 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 8 |
Hb_001558_070 |
0.1903399307 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 9 |
Hb_000429_110 |
0.1906009165 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 10 |
Hb_009736_010 |
0.1924657729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000111_480 |
0.1926151666 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 12 |
Hb_000977_090 |
0.1952989684 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 13 |
Hb_001355_070 |
0.1978090659 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein, putative isoform 2 [Theobroma cacao] |
| 14 |
Hb_002928_180 |
0.2014755991 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 15 |
Hb_000059_300 |
0.2027439284 |
- |
- |
caspase, putative [Ricinus communis] |
| 16 |
Hb_006573_300 |
0.2068905584 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_001959_120 |
0.2084879732 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 18 |
Hb_001049_160 |
0.2094890591 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 19 |
Hb_000685_060 |
0.2095478446 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_000042_250 |
0.2109646373 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |