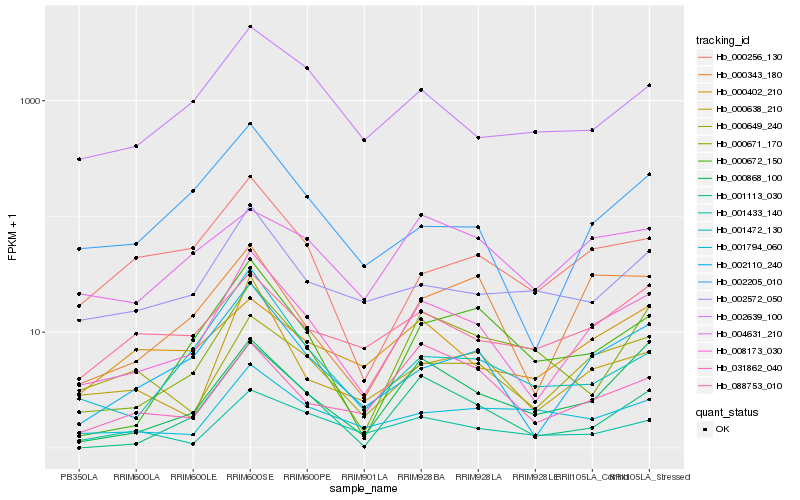
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008173_030 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 2 |
Hb_002110_240 |
0.1360808631 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 3 |
Hb_000649_240 |
0.1698618228 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 4 |
Hb_000868_100 |
0.1722327547 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 5 |
Hb_000343_180 |
0.1760430659 |
- |
- |
NDWp3 [Podospora anserina] |
| 6 |
Hb_002205_010 |
0.1785750191 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 7 |
Hb_000638_210 |
0.1812859083 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_002639_100 |
0.1836714188 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 9 |
Hb_001113_030 |
0.1839435015 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 10 |
Hb_000672_150 |
0.1877646451 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 11 |
Hb_001433_140 |
0.1905049736 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 12 |
Hb_002572_050 |
0.191552035 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000256_130 |
0.1917203452 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 14 |
Hb_088753_010 |
0.1936863893 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 15 |
Hb_001472_130 |
0.1965128535 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 16 |
Hb_001794_060 |
0.1971952038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000671_170 |
0.1983017337 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_004631_210 |
0.2005626896 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 19 |
Hb_031862_040 |
0.2009076455 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 20 |
Hb_000402_210 |
0.2026724534 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |