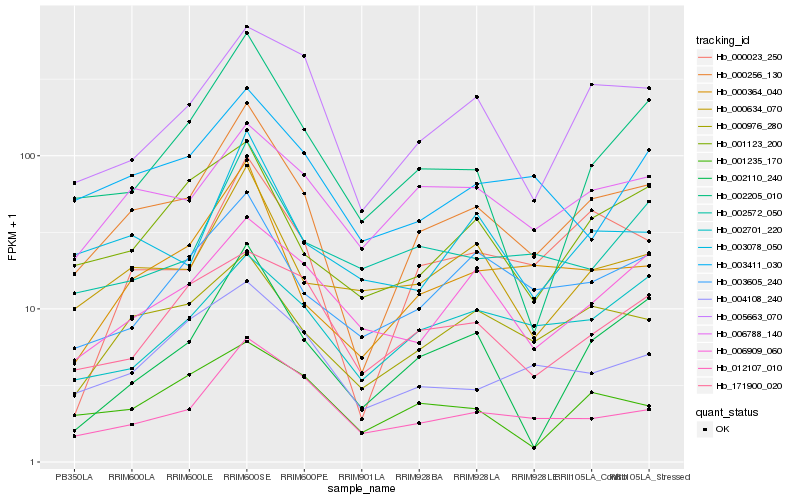
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_130 |
0.0 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 2 |
Hb_000023_250 |
0.1415318706 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 3 |
Hb_000364_040 |
0.143162637 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 4 |
Hb_000976_280 |
0.1441845258 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 5 |
Hb_012107_010 |
0.1547782749 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 6 |
Hb_000634_070 |
0.1557219848 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 7 |
Hb_005663_070 |
0.1582156857 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 8 |
Hb_003605_240 |
0.1591137642 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 9 |
Hb_003078_050 |
0.1604225055 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 10 |
Hb_006788_140 |
0.1615266746 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 11 |
Hb_002205_010 |
0.1657254687 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 12 |
Hb_002110_240 |
0.1664296024 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 13 |
Hb_001123_200 |
0.1665400267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004108_240 |
0.1716643675 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 15 |
Hb_006909_060 |
0.1755824818 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 16 |
Hb_003411_030 |
0.1773005092 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |
| 17 |
Hb_002572_050 |
0.1788593297 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001235_170 |
0.1816534348 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 19 |
Hb_002701_220 |
0.1820403711 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 20 |
Hb_171900_020 |
0.1836135544 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |