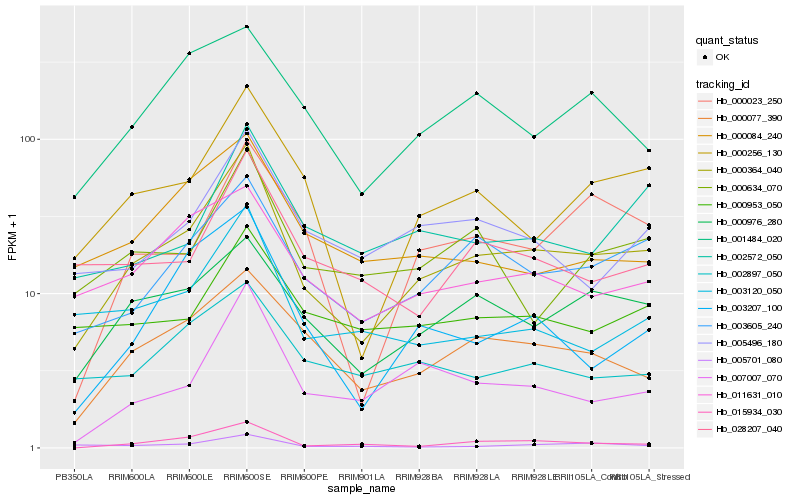
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000364_040 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 2 |
Hb_003605_240 |
0.1383650475 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 3 |
Hb_000256_130 |
0.143162637 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 4 |
Hb_000976_280 |
0.1485060427 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 5 |
Hb_000077_390 |
0.1535293321 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0019s13330g [Populus trichocarpa] |
| 6 |
Hb_015934_030 |
0.1615306671 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_011631_010 |
0.1655741936 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 8 |
Hb_005496_180 |
0.1674651318 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 9 |
Hb_007007_070 |
0.1682509092 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 10 |
Hb_003207_100 |
0.1708857435 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 11 |
Hb_000634_070 |
0.1735656607 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 12 |
Hb_001484_020 |
0.1745239503 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_003120_050 |
0.1752955673 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 14 |
Hb_028207_040 |
0.1755297332 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 15 |
Hb_000023_250 |
0.1770438802 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 16 |
Hb_002572_050 |
0.1778529704 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002897_050 |
0.178746167 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 18 |
Hb_000953_050 |
0.1790098212 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 19 |
Hb_000084_240 |
0.1813045471 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 20 |
Hb_005701_080 |
0.1816414587 |
- |
- |
ATP binding protein, putative [Ricinus communis] |