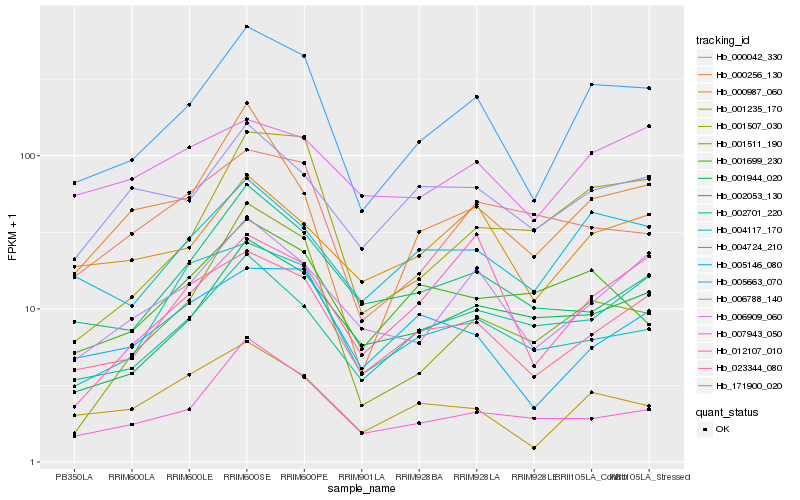
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005663_070 |
0.0 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 2 |
Hb_171900_020 |
0.1277912737 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 3 |
Hb_006909_060 |
0.1354910173 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 4 |
Hb_004724_210 |
0.1381094128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001944_020 |
0.1489117515 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 6 |
Hb_001235_170 |
0.151061495 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 7 |
Hb_012107_010 |
0.1533937017 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 8 |
Hb_001507_030 |
0.156420302 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 9 |
Hb_000987_060 |
0.1581273002 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 10 |
Hb_000256_130 |
0.1582156857 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 11 |
Hb_002053_130 |
0.1660801378 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 12 |
Hb_005146_080 |
0.1672887865 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006788_140 |
0.1679103072 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 14 |
Hb_002701_220 |
0.1697039062 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 15 |
Hb_001511_190 |
0.1710793075 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 16 |
Hb_023344_080 |
0.1774770757 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001699_230 |
0.1781856476 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_000042_330 |
0.1784143442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007943_050 |
0.1793067044 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 20 |
Hb_004117_170 |
0.1806459227 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |