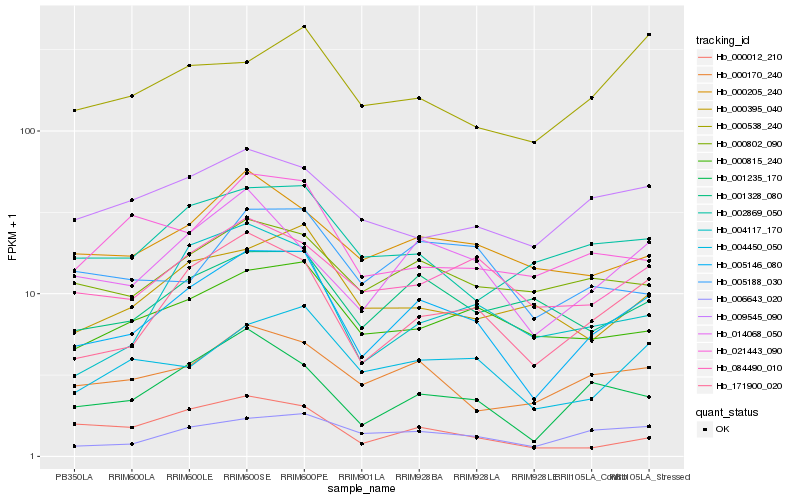
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005146_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 2 |
Hb_171900_020 |
0.1013487644 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 3 |
Hb_004450_050 |
0.126554126 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 4 |
Hb_001235_170 |
0.1301914933 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 5 |
Hb_006643_020 |
0.1345836604 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 6 |
Hb_014068_050 |
0.1371730353 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 7 |
Hb_000170_240 |
0.142044652 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 8 |
Hb_084490_010 |
0.1470836455 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000815_240 |
0.1486723351 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000802_090 |
0.1528688661 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_000205_240 |
0.1535918048 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_001328_080 |
0.1553028801 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 13 |
Hb_002869_050 |
0.1569951722 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000395_040 |
0.1572606244 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 15 |
Hb_005188_030 |
0.1578274402 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_004117_170 |
0.1614204073 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 17 |
Hb_009545_090 |
0.1626455486 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 18 |
Hb_000012_210 |
0.1626960209 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_021443_090 |
0.1629313862 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 20 |
Hb_000538_240 |
0.164799295 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |