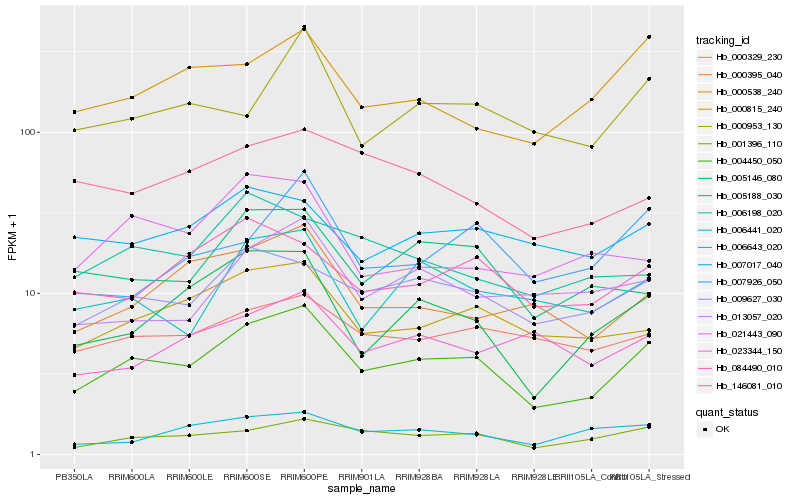
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004450_050 |
0.0 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 2 |
Hb_001396_110 |
0.1164550658 |
- |
- |
DNA primase large subunit, putative [Ricinus communis] |
| 3 |
Hb_009627_030 |
0.1208431924 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 4 |
Hb_000815_240 |
0.1231565352 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005146_080 |
0.126554126 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005188_030 |
0.1400916911 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_006441_020 |
0.1409960884 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 8 |
Hb_000395_040 |
0.1411735297 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 9 |
Hb_000329_230 |
0.1416487826 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 10 |
Hb_000538_240 |
0.1441118406 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 11 |
Hb_084490_010 |
0.1443573246 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000953_130 |
0.1456660055 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 13 |
Hb_006643_020 |
0.1474816083 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 14 |
Hb_013057_020 |
0.1485773151 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 15 |
Hb_007017_040 |
0.1491353083 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 16 |
Hb_146081_010 |
0.1518264281 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 17 |
Hb_007926_050 |
0.1533548017 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 18 |
Hb_006198_020 |
0.1557478528 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 19 |
Hb_023344_150 |
0.1557623939 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 20 |
Hb_021443_090 |
0.156771456 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |