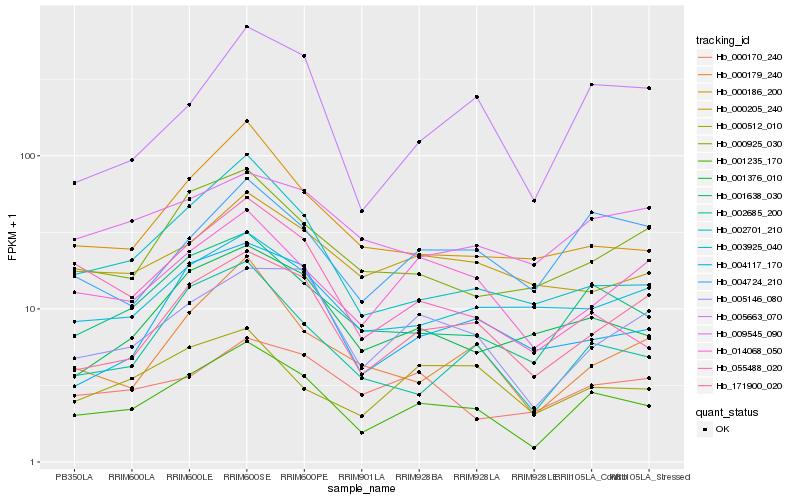
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_170 |
0.0 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 2 |
Hb_001638_030 |
0.1026710821 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 3 |
Hb_171900_020 |
0.1242185764 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 4 |
Hb_005146_080 |
0.1301914933 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002685_200 |
0.1310226845 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 6 |
Hb_014068_050 |
0.1357462699 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 7 |
Hb_001376_010 |
0.1442140851 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 8 |
Hb_000925_030 |
0.1491330589 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_004724_210 |
0.1494311045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002701_210 |
0.1503810554 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 11 |
Hb_000179_240 |
0.1506307549 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005663_070 |
0.151061495 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 13 |
Hb_003925_040 |
0.1538000276 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 14 |
Hb_000186_200 |
0.1544111696 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 15 |
Hb_055488_020 |
0.1557053061 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 16 |
Hb_004117_170 |
0.1561242523 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 17 |
Hb_000512_010 |
0.1573113033 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 18 |
Hb_009545_090 |
0.1574524777 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 19 |
Hb_000205_240 |
0.1586090976 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000170_240 |
0.1601549901 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |