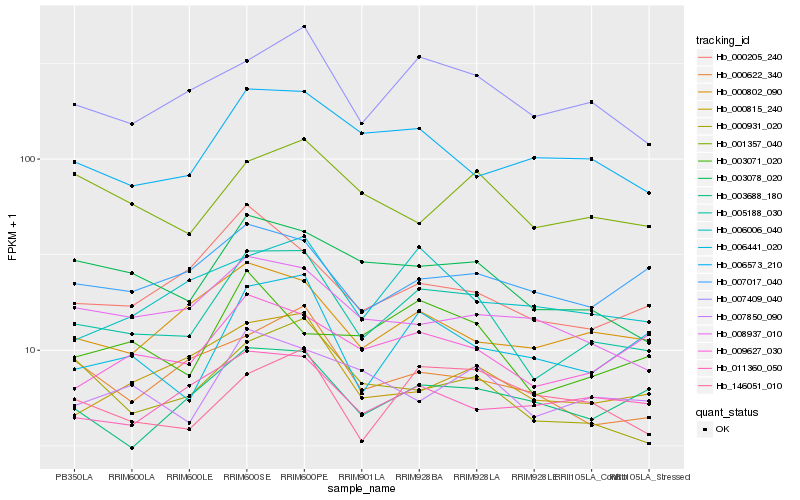
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005188_030 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_007409_040 |
0.100974456 |
- |
- |
actin family protein [Populus trichocarpa] |
| 3 |
Hb_003078_020 |
0.1080563265 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 4 |
Hb_000931_020 |
0.1154342821 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000802_090 |
0.120666935 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_000815_240 |
0.122670292 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_006441_020 |
0.1228337235 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 8 |
Hb_000205_240 |
0.1234050946 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_009627_030 |
0.1255715816 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 10 |
Hb_006573_210 |
0.1256728673 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 11 |
Hb_007850_090 |
0.1268133275 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000622_340 |
0.1271402828 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 13 |
Hb_003688_180 |
0.1279443256 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 14 |
Hb_008937_010 |
0.1302944186 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006006_040 |
0.1305808835 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 16 |
Hb_003071_020 |
0.1328248532 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 17 |
Hb_007017_040 |
0.1328501132 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 18 |
Hb_011360_050 |
0.1338107605 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001357_040 |
0.1340740163 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 20 |
Hb_146051_010 |
0.1347451443 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |