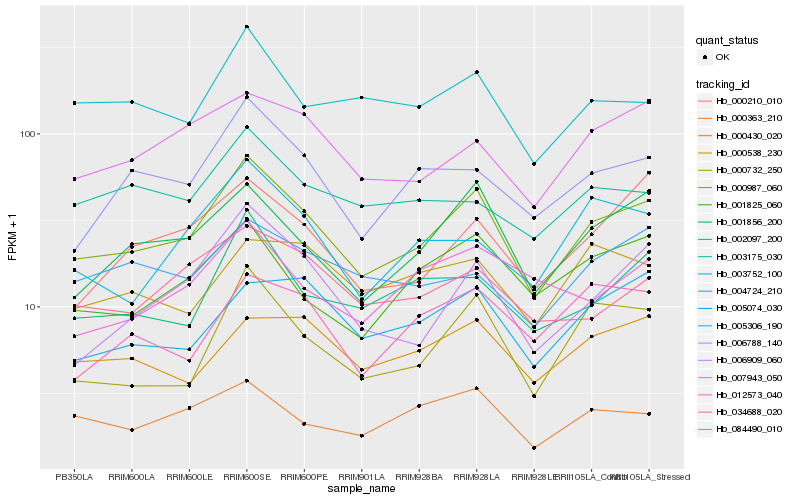
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000987_060 |
0.0 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 2 |
Hb_001856_200 |
0.1016307886 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 3 |
Hb_000732_250 |
0.1127384473 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 4 |
Hb_006788_140 |
0.1128117797 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 5 |
Hb_007943_050 |
0.1159372475 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 6 |
Hb_006909_060 |
0.1160427694 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 7 |
Hb_002097_200 |
0.1167321703 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_001825_060 |
0.1197362466 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 9 |
Hb_003752_100 |
0.1215619101 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 10 |
Hb_000363_210 |
0.1222418721 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 11 |
Hb_084490_010 |
0.1244479659 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000210_010 |
0.1259882304 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 13 |
Hb_000430_020 |
0.1269745347 |
- |
- |
- |
| 14 |
Hb_004724_210 |
0.1277608654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005074_030 |
0.1284315957 |
- |
- |
- |
| 16 |
Hb_005306_190 |
0.1311777604 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 17 |
Hb_034688_020 |
0.1343975277 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 18 |
Hb_003175_030 |
0.1347319529 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 19 |
Hb_012573_040 |
0.1364897658 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 20 |
Hb_000538_230 |
0.1379636512 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |