| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
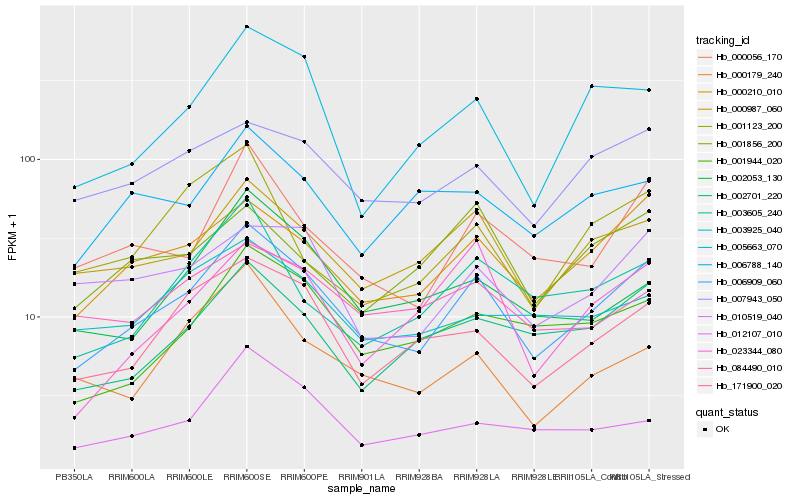
Hb_006909_060 |
0.0 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 2 |
Hb_000210_010 |
0.1102410823 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 3 |
Hb_000987_060 |
0.1160427694 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 4 |
Hb_000056_170 |
0.1243939183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_171900_020 |
0.127039128 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 6 |
Hb_002701_220 |
0.1309369333 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 7 |
Hb_001944_020 |
0.1339219352 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 8 |
Hb_005663_070 |
0.1354910173 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 9 |
Hb_007943_050 |
0.1357229322 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 10 |
Hb_003605_240 |
0.1404353504 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 11 |
Hb_023344_080 |
0.1475893852 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010519_040 |
0.1476856311 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 13 |
Hb_006788_140 |
0.1481778648 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 14 |
Hb_002053_130 |
0.1488959382 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 15 |
Hb_084490_010 |
0.1557527542 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000179_240 |
0.1558263017 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003925_040 |
0.1564816955 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 18 |
Hb_012107_010 |
0.1583260736 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 19 |
Hb_001856_200 |
0.1585256724 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 20 |
Hb_001123_200 |
0.1599998761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |