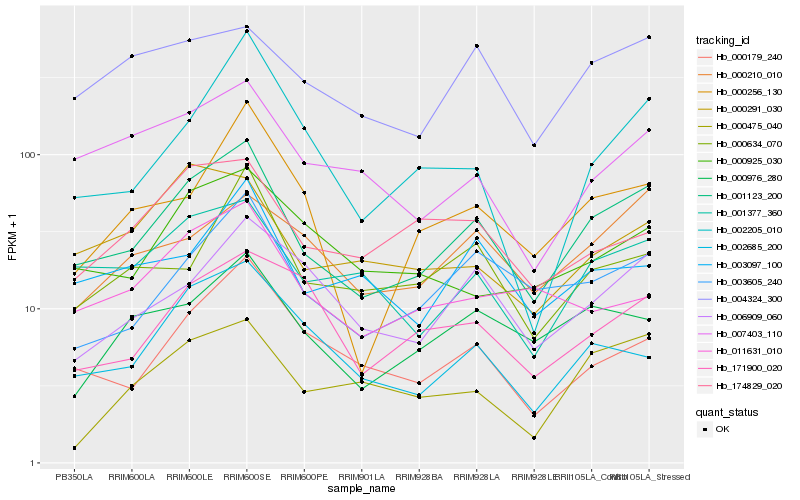
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003605_240 |
0.1363321663 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 3 |
Hb_001377_360 |
0.1408737323 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 4 |
Hb_000179_240 |
0.1511392037 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000976_280 |
0.155915722 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 6 |
Hb_002685_200 |
0.159598157 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 7 |
Hb_004324_300 |
0.1597025951 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 8 |
Hb_006909_060 |
0.1599998761 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 9 |
Hb_007403_110 |
0.1604795667 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 10 |
Hb_003097_100 |
0.1644142125 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 11 |
Hb_000634_070 |
0.1644178297 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 12 |
Hb_000256_130 |
0.1665400267 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 13 |
Hb_000210_010 |
0.1673731493 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 14 |
Hb_000475_040 |
0.1681243433 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000925_030 |
0.1703912439 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_174829_020 |
0.1746952857 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 17 |
Hb_000291_030 |
0.1749856111 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 18 |
Hb_171900_020 |
0.175078921 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 19 |
Hb_002205_010 |
0.1750974669 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 20 |
Hb_011631_010 |
0.1760776411 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |