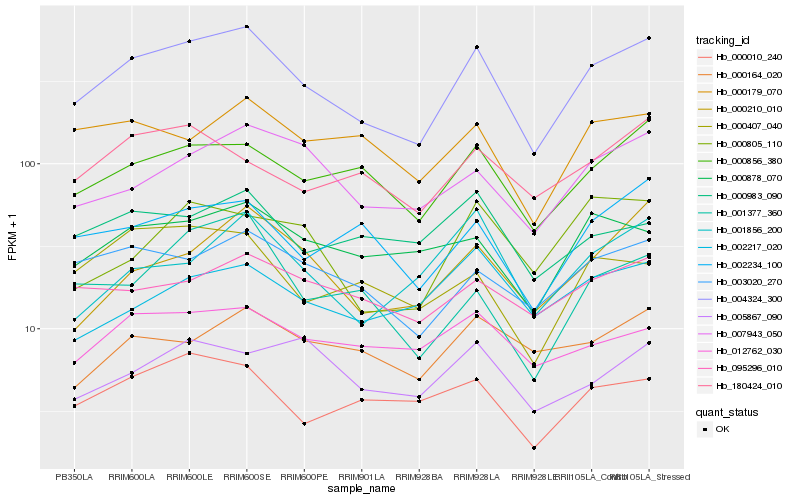
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_300 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 2 |
Hb_000856_380 |
0.1038387388 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 3 |
Hb_003020_270 |
0.1143399909 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 4 |
Hb_007943_050 |
0.1181901673 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 5 |
Hb_002234_100 |
0.1190641422 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 6 |
Hb_000210_010 |
0.1248039908 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 7 |
Hb_000010_240 |
0.1253456638 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 8 |
Hb_000878_070 |
0.1272540083 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 9 |
Hb_012762_030 |
0.1287277458 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 10 |
Hb_000179_070 |
0.1296484205 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 11 |
Hb_095296_010 |
0.1298562033 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 12 |
Hb_001377_360 |
0.1329940623 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 13 |
Hb_001856_200 |
0.1336948888 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 14 |
Hb_180424_010 |
0.1338803533 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 15 |
Hb_000164_020 |
0.1346610807 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 16 |
Hb_000983_090 |
0.1348649699 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 17 |
Hb_000407_040 |
0.1357020884 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000805_110 |
0.1363197513 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 19 |
Hb_002217_020 |
0.1396802171 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005867_090 |
0.1402262098 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |