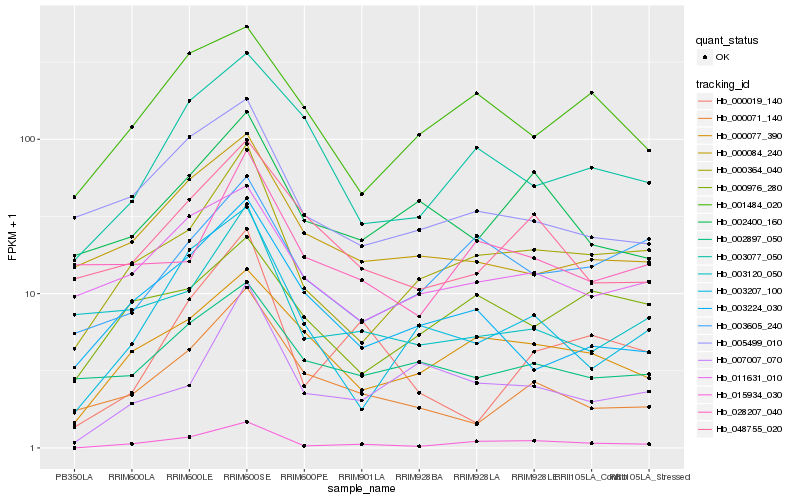
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015934_030 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000364_040 |
0.1615306671 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 3 |
Hb_000077_390 |
0.1918121338 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0019s13330g [Populus trichocarpa] |
| 4 |
Hb_003207_100 |
0.2113392475 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 5 |
Hb_003605_240 |
0.2172536322 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 6 |
Hb_003077_050 |
0.2201336861 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 7 |
Hb_007007_070 |
0.2218697528 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 8 |
Hb_000019_140 |
0.2225507112 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 9 |
Hb_001484_020 |
0.2250299924 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_005499_010 |
0.2260152958 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 11 |
Hb_011631_010 |
0.2277465433 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 12 |
Hb_048755_020 |
0.2286362089 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 13 |
Hb_002400_160 |
0.2293097858 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 14 |
Hb_000071_140 |
0.2318188677 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 15 |
Hb_000084_240 |
0.2322382576 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 16 |
Hb_000976_280 |
0.2358184358 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 17 |
Hb_028207_040 |
0.2368231101 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 18 |
Hb_003120_050 |
0.2376600014 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 19 |
Hb_003224_030 |
0.2387171098 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 20 |
Hb_002897_050 |
0.2388837747 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |