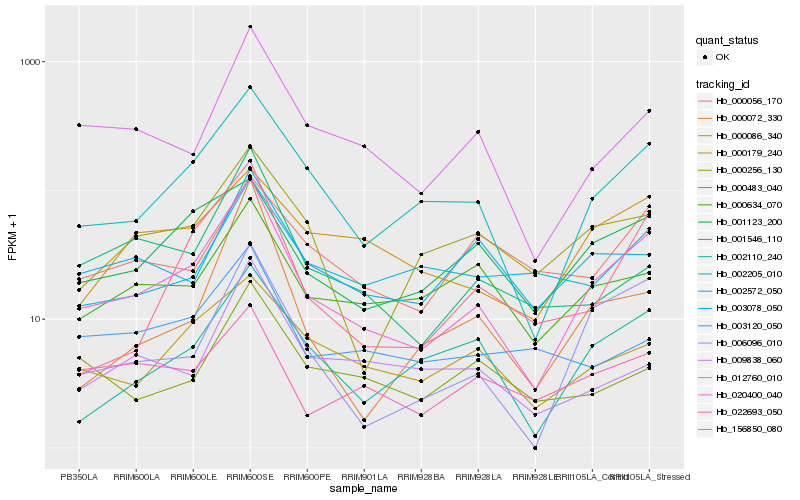
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020400_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 2 |
Hb_002205_010 |
0.1221242064 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 3 |
Hb_006096_010 |
0.1617175571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012760_010 |
0.1668574136 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002110_240 |
0.1707302415 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 6 |
Hb_156850_080 |
0.1768311557 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 7 |
Hb_001123_200 |
0.1820089829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000179_240 |
0.1923840003 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000634_070 |
0.1965233126 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 10 |
Hb_000256_130 |
0.1966142946 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 11 |
Hb_001546_110 |
0.1970722505 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 3 [Jatropha curcas] |
| 12 |
Hb_022693_050 |
0.2005429182 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 13 |
Hb_003078_050 |
0.2007690145 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 14 |
Hb_000086_340 |
0.201721906 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 15 |
Hb_002572_050 |
0.2130965293 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000056_170 |
0.2142885206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009838_060 |
0.2157850522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003120_050 |
0.2159070973 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 19 |
Hb_000072_330 |
0.2186568116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000483_040 |
0.2195517246 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |