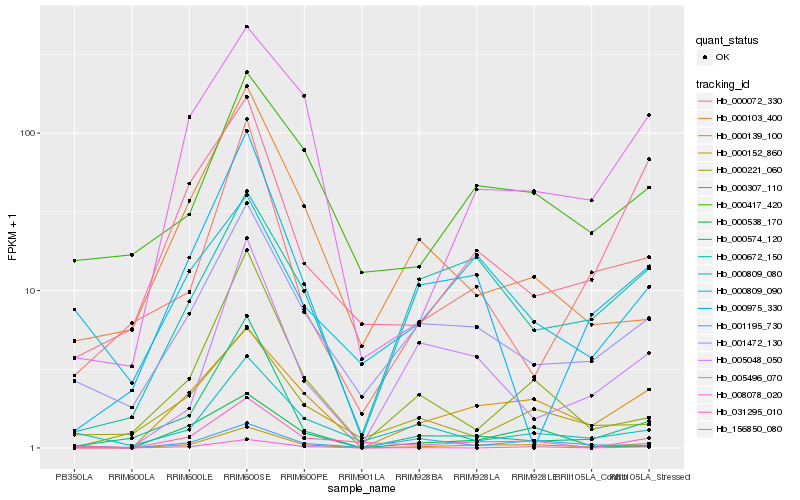
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_860 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000139_100 |
0.168627162 |
- |
- |
PREDICTED: sugar transport protein 13-like [Jatropha curcas] |
| 3 |
Hb_001472_130 |
0.1761460657 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 4 |
Hb_031295_010 |
0.1956940739 |
- |
- |
hypothetical protein JCGZ_19530 [Jatropha curcas] |
| 5 |
Hb_000417_420 |
0.2053219536 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 6 |
Hb_000809_080 |
0.2065881144 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 7 |
Hb_005496_070 |
0.2132375819 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 8 |
Hb_000221_060 |
0.2234812553 |
- |
- |
- |
| 9 |
Hb_156850_080 |
0.2278295092 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 10 |
Hb_000809_090 |
0.233614766 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 11 |
Hb_000672_150 |
0.2384339574 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 12 |
Hb_000072_330 |
0.2400793887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000538_170 |
0.2436734154 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_000975_330 |
0.244280939 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 15 |
Hb_000574_120 |
0.2454646511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005048_050 |
0.2483104768 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_000103_400 |
0.2485830473 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000307_110 |
0.249893734 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 19 |
Hb_001195_730 |
0.253492774 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 20 |
Hb_008078_020 |
0.2535284898 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |