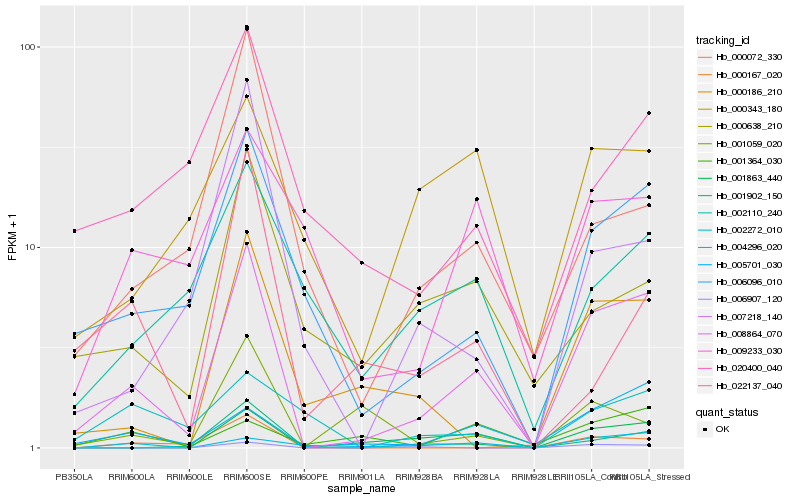
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105803851 [Gossypium raimondii] |
| 2 |
Hb_006096_010 |
0.2260564983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005701_030 |
0.253904176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001863_440 |
0.2579714468 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 5 |
Hb_001902_150 |
0.2656186772 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |
| 6 |
Hb_000186_210 |
0.2790827693 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 7 |
Hb_000167_020 |
0.2863627372 |
- |
- |
- |
| 8 |
Hb_004296_020 |
0.2943371349 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_006907_120 |
0.3020227883 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 10 |
Hb_007218_140 |
0.3107590541 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 11 |
Hb_020400_040 |
0.3126228159 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 12 |
Hb_000072_330 |
0.3126900168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002110_240 |
0.3135638937 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_009233_030 |
0.3153598072 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000638_210 |
0.3157918596 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_001059_020 |
0.319167537 |
- |
- |
PREDICTED: FBD-associated F-box protein At4g10400-like [Malus domestica] |
| 17 |
Hb_000343_180 |
0.3232318675 |
- |
- |
NDWp3 [Podospora anserina] |
| 18 |
Hb_001364_030 |
0.3264935597 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 19 |
Hb_002272_010 |
0.3312034754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_022137_040 |
0.331553478 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |