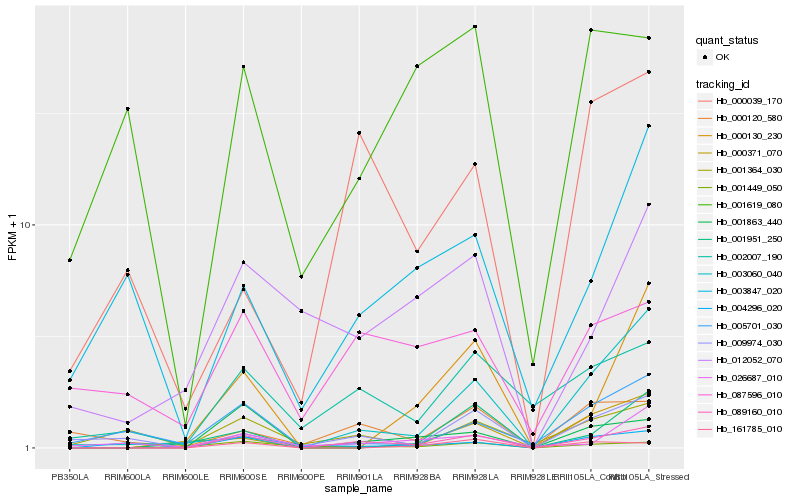
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089160_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 2 |
Hb_001863_440 |
0.2209138869 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 3 |
Hb_001449_050 |
0.2552964672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002007_190 |
0.2696428248 |
- |
- |
hypothetical protein RCOM_1432220 [Ricinus communis] |
| 5 |
Hb_009974_030 |
0.2811741199 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_161785_010 |
0.291977586 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 7 |
Hb_000130_230 |
0.2957121808 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 8 |
Hb_000120_580 |
0.2985320505 |
- |
- |
hypothetical protein POPTR_0007s08570g [Populus trichocarpa] |
| 9 |
Hb_012052_070 |
0.302104265 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 10 |
Hb_001951_250 |
0.3039263183 |
- |
- |
- |
| 11 |
Hb_001364_030 |
0.3039407464 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 12 |
Hb_087596_010 |
0.3043087817 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 13 |
Hb_000039_170 |
0.3122512782 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 14 |
Hb_004296_020 |
0.3131383405 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_005701_030 |
0.3151499457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003060_040 |
0.3176272763 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 17 |
Hb_026687_010 |
0.3242849119 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 18 |
Hb_003847_020 |
0.3255816926 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_001619_080 |
0.3266451793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000371_070 |
0.3308998777 |
- |
- |
PREDICTED: uncharacterized protein LOC103863319 [Brassica rapa] |