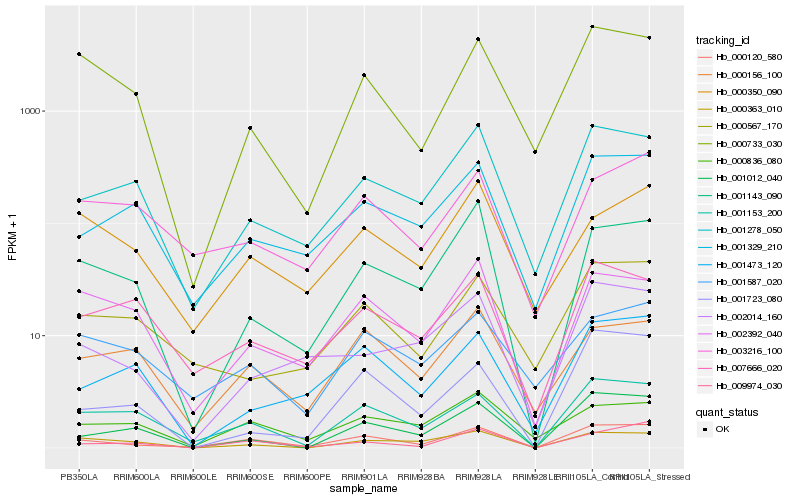
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_580 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s08570g [Populus trichocarpa] |
| 2 |
Hb_001153_200 |
0.1466476035 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009974_030 |
0.1634185637 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001278_050 |
0.1733759974 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 5 |
Hb_001143_090 |
0.1746032845 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 6 [Jatropha curcas] |
| 6 |
Hb_001329_210 |
0.1760097791 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_001012_040 |
0.1852140054 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_002392_040 |
0.1877491636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002014_160 |
0.1916855097 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor C-1 [Jatropha curcas] |
| 10 |
Hb_000156_100 |
0.1927165894 |
- |
- |
PREDICTED: uncharacterized mitochondrial protein ymf11 [Jatropha curcas] |
| 11 |
Hb_000733_030 |
0.1928533106 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 12 |
Hb_000836_080 |
0.1954947401 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g46050, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001473_120 |
0.1960645146 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_001723_080 |
0.1964215922 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 15 |
Hb_007666_020 |
0.1976666076 |
- |
- |
alkaline phytoceramidase, putative [Ricinus communis] |
| 16 |
Hb_003216_100 |
0.1985815227 |
- |
- |
PREDICTED: uncharacterized protein LOC105641496 [Jatropha curcas] |
| 17 |
Hb_001587_020 |
0.1995102596 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 18 |
Hb_000350_090 |
0.2006828111 |
transcription factor |
TF Family: GeBP |
transcription regulator, putative [Ricinus communis] |
| 19 |
Hb_000363_010 |
0.2012947575 |
- |
- |
- |
| 20 |
Hb_000567_170 |
0.2022591128 |
- |
- |
PREDICTED: putative lipase YDR444W isoform X1 [Jatropha curcas] |