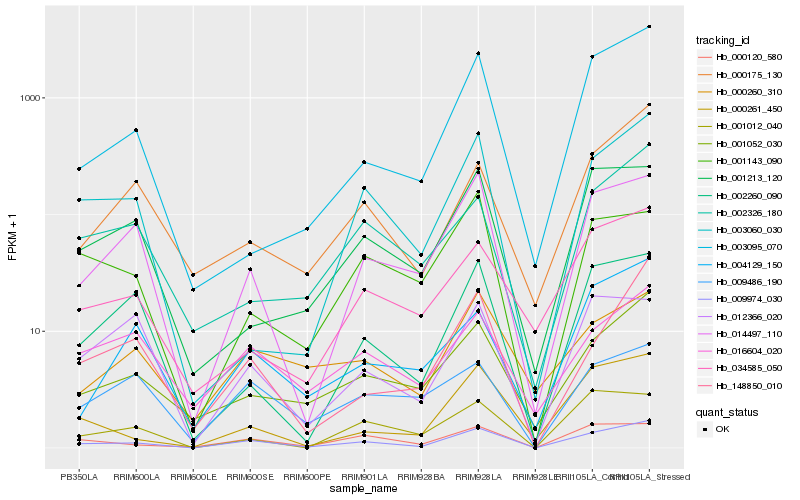
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009974_030 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_014497_110 |
0.1605378654 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 3 |
Hb_000120_580 |
0.1634185637 |
- |
- |
hypothetical protein POPTR_0007s08570g [Populus trichocarpa] |
| 4 |
Hb_001052_030 |
0.1712064294 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000261_450 |
0.1899339773 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 6 |
Hb_016604_020 |
0.1944334937 |
- |
- |
- |
| 7 |
Hb_003060_030 |
0.1950797658 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 8 |
Hb_148850_010 |
0.1956660622 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 9 |
Hb_002260_090 |
0.1959761506 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000260_310 |
0.1963678641 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 11 |
Hb_000175_130 |
0.1978067857 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 12 |
Hb_001012_040 |
0.1980621562 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_009486_190 |
0.1995198422 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_004129_150 |
0.1998636996 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 15 |
Hb_002326_180 |
0.200763363 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 16 |
Hb_034585_050 |
0.2024536746 |
- |
- |
- |
| 17 |
Hb_001213_120 |
0.2024961569 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 18 |
Hb_001143_090 |
0.2043141073 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 6 [Jatropha curcas] |
| 19 |
Hb_003095_070 |
0.2052338552 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 20 |
Hb_012366_020 |
0.2055702089 |
- |
- |
unnamed protein product [Vitis vinifera] |