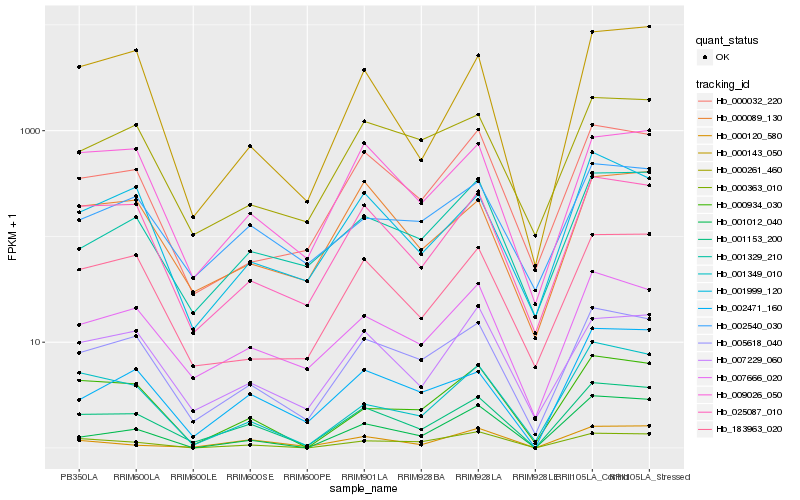
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_200 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005618_040 |
0.1023135964 |
- |
- |
- |
| 3 |
Hb_007666_020 |
0.1258296711 |
- |
- |
alkaline phytoceramidase, putative [Ricinus communis] |
| 4 |
Hb_025087_010 |
0.1266568102 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 5 |
Hb_009026_050 |
0.1386132338 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 6 |
Hb_002471_160 |
0.1395728325 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4-like [Populus euphratica] |
| 7 |
Hb_000934_030 |
0.1439298784 |
- |
- |
- |
| 8 |
Hb_000120_580 |
0.1466476035 |
- |
- |
hypothetical protein POPTR_0007s08570g [Populus trichocarpa] |
| 9 |
Hb_001999_120 |
0.1476139752 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_000089_130 |
0.1480568889 |
- |
- |
mitochondrial ribosomal protein S16, putative [Ricinus communis] |
| 11 |
Hb_000143_050 |
0.1484300558 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 12 |
Hb_001349_010 |
0.1489331404 |
- |
- |
- |
| 13 |
Hb_183963_020 |
0.1490068082 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 14 |
Hb_001329_210 |
0.149069957 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 15 |
Hb_007229_060 |
0.1500088527 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g44880 [Jatropha curcas] |
| 16 |
Hb_002540_030 |
0.1507300427 |
- |
- |
hypothetical protein 32 [Hevea brasiliensis] |
| 17 |
Hb_000032_220 |
0.1508134933 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000363_010 |
0.1509452844 |
- |
- |
- |
| 19 |
Hb_000261_460 |
0.1526497808 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 20 |
Hb_001012_040 |
0.1533773687 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |