| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
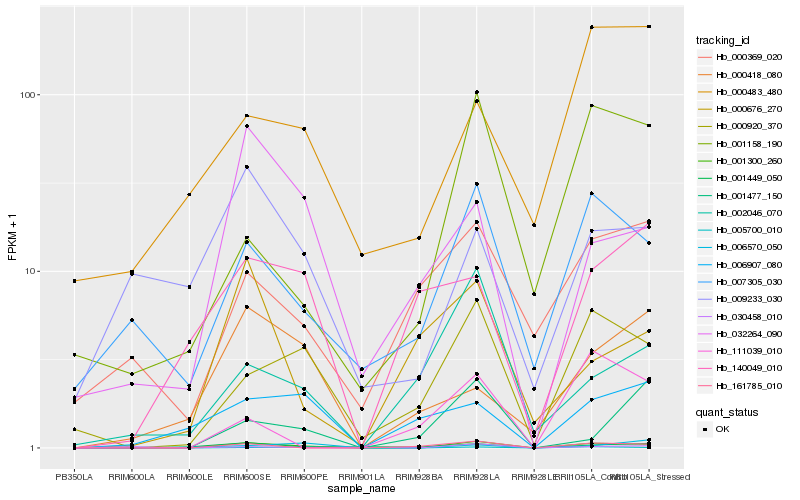
Hb_001300_260 |
0.0 |
- |
- |
- |
| 2 |
Hb_000920_370 |
0.2556067601 |
- |
- |
hypothetical protein JCGZ_24527 [Jatropha curcas] |
| 3 |
Hb_161785_010 |
0.2823224214 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 4 |
Hb_007305_030 |
0.288037368 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001449_050 |
0.3017523388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001158_190 |
0.3208544671 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 7 |
Hb_032264_090 |
0.3329001222 |
- |
- |
hypothetical protein JCGZ_02512 [Jatropha curcas] |
| 8 |
Hb_030458_010 |
0.3380055784 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_002046_070 |
0.3417492566 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 10 |
Hb_006907_080 |
0.3441895329 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 11 |
Hb_005700_010 |
0.3461054013 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000369_020 |
0.3465371354 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 13 |
Hb_009233_030 |
0.3487526928 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_111039_010 |
0.3504844315 |
- |
- |
- |
| 15 |
Hb_001477_150 |
0.3506030677 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000676_270 |
0.3518494587 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 17 |
Hb_000418_080 |
0.3535988331 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 18 |
Hb_140049_010 |
0.35410344 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 19 |
Hb_006570_050 |
0.3566139633 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 20 |
Hb_000483_480 |
0.357843884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |