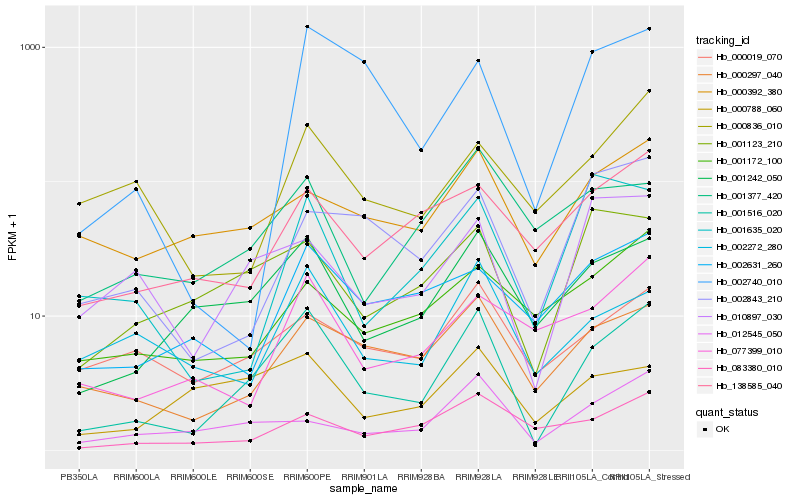
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001516_020 |
0.0 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-9-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001242_050 |
0.1910595487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000297_040 |
0.2102475599 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 4 |
Hb_001123_210 |
0.2316028157 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 5 |
Hb_002272_280 |
0.2328139267 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 6 |
Hb_012545_050 |
0.2334652271 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 7 |
Hb_083380_010 |
0.2344860064 |
- |
- |
- |
| 8 |
Hb_010897_030 |
0.2361645778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000019_070 |
0.2386657646 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 10 |
Hb_077399_010 |
0.2408589641 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 11 |
Hb_000392_380 |
0.2417086264 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 12 |
Hb_138585_040 |
0.2420812255 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 13 |
Hb_000788_060 |
0.2464570318 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 14 |
Hb_002740_010 |
0.2491634001 |
- |
- |
stress-induced hydrophobic peptide [Cleistogenes songorica] |
| 15 |
Hb_002631_260 |
0.2492538912 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 16 |
Hb_001377_420 |
0.2506453478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001635_020 |
0.2513819587 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 18 |
Hb_002843_210 |
0.252173754 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 19 |
Hb_000836_010 |
0.2524534086 |
- |
- |
beta-cyanoalanine synthase [Hevea brasiliensis] |
| 20 |
Hb_001172_100 |
0.2535064658 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |