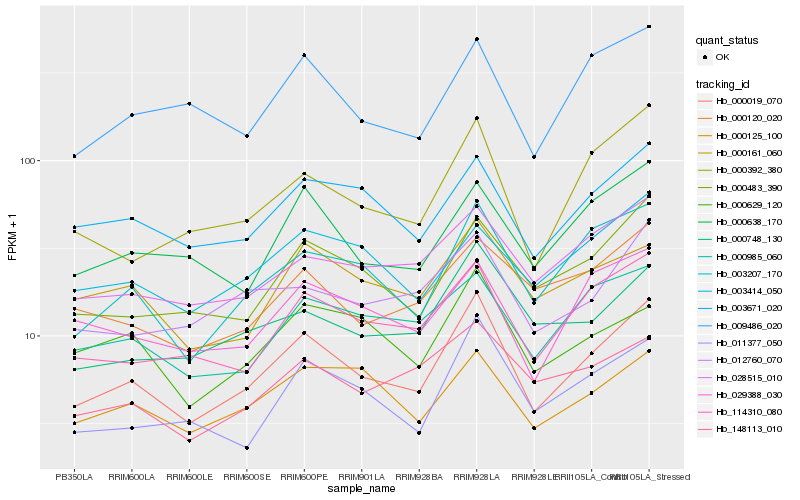
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 2 |
Hb_003207_170 |
0.0896184262 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 3 |
Hb_114310_080 |
0.0973111279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000483_390 |
0.1025903939 |
- |
- |
PREDICTED: vesicle-associated protein 2-1 [Jatropha curcas] |
| 5 |
Hb_011377_050 |
0.1035224348 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000392_380 |
0.1039189887 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 7 |
Hb_003671_020 |
0.1078822483 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 8 |
Hb_000748_130 |
0.1129652674 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000638_170 |
0.1132617349 |
- |
- |
PREDICTED: ras-related protein RABC1 [Jatropha curcas] |
| 10 |
Hb_028515_010 |
0.1142346973 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029388_030 |
0.1144834211 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 12 |
Hb_000985_060 |
0.1157328977 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_009486_020 |
0.1159416291 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 14 |
Hb_012760_070 |
0.1189944152 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_000161_060 |
0.1211687348 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 [Jatropha curcas] |
| 16 |
Hb_000125_100 |
0.1212143945 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105635070 [Jatropha curcas] |
| 17 |
Hb_000120_020 |
0.1225316256 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 18 |
Hb_000629_120 |
0.1237047837 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |
| 19 |
Hb_148113_010 |
0.1239578708 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17C-like isoform X3 [Citrus sinensis] |
| 20 |
Hb_003414_050 |
0.1246614008 |
- |
- |
PREDICTED: uncharacterized protein LOC105640180 isoform X3 [Jatropha curcas] |