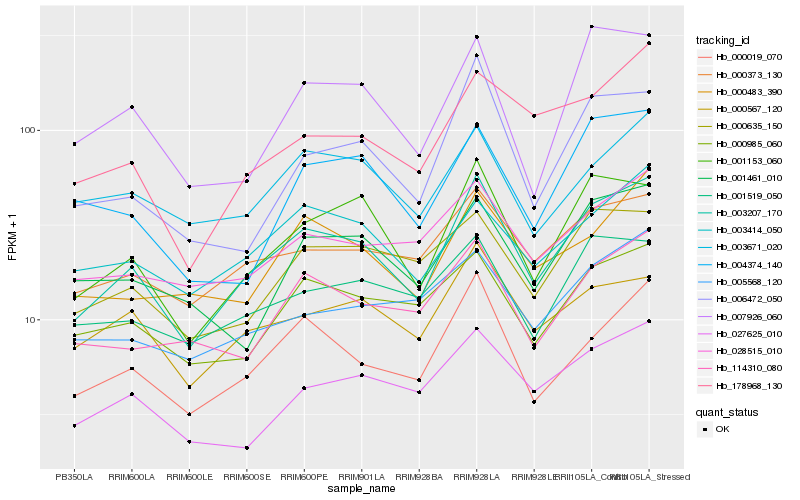
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_170 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001153_060 |
0.0860156113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000019_070 |
0.0896184262 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 4 |
Hb_000373_130 |
0.1026172191 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 5 |
Hb_000567_120 |
0.1087867146 |
- |
- |
PREDICTED: transcription factor IIIB 50 kDa subunit-like [Jatropha curcas] |
| 6 |
Hb_000985_060 |
0.1092233497 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_027625_010 |
0.109315117 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 8 |
Hb_114310_080 |
0.1098830875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_178968_130 |
0.1107064236 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 10 |
Hb_028515_010 |
0.1111292711 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000483_390 |
0.1118295406 |
- |
- |
PREDICTED: vesicle-associated protein 2-1 [Jatropha curcas] |
| 12 |
Hb_003414_050 |
0.1118964819 |
- |
- |
PREDICTED: uncharacterized protein LOC105640180 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000635_150 |
0.1132781375 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 14 |
Hb_006472_050 |
0.1133855909 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_001519_050 |
0.1145875248 |
- |
- |
PREDICTED: protein RMD5 homolog A-like [Jatropha curcas] |
| 16 |
Hb_007926_060 |
0.1153223705 |
- |
- |
ubiquitin-conjugating enzyme E2 variant 1D [Arabidopsis thaliana] |
| 17 |
Hb_001461_010 |
0.1175890149 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Pyrus x bretschneideri] |
| 18 |
Hb_003671_020 |
0.1193420942 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 19 |
Hb_005568_120 |
0.11951636 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 20 |
Hb_004374_140 |
0.1204887836 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |