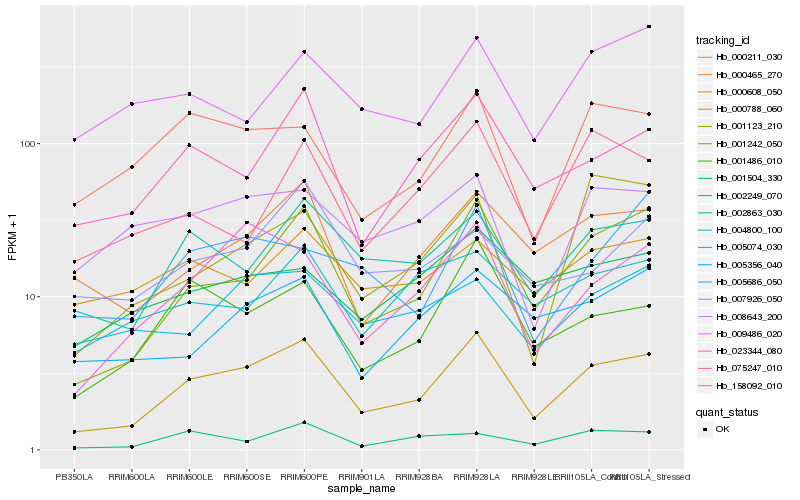
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000788_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 2 |
Hb_001242_050 |
0.1072775936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_075247_010 |
0.1393522014 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 4 |
Hb_000465_270 |
0.1432646168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_023344_080 |
0.1439189288 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002863_030 |
0.1444490279 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 7 |
Hb_001486_010 |
0.1512206309 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 8 |
Hb_000211_030 |
0.1637552409 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 9 |
Hb_005356_040 |
0.1680290856 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_008643_200 |
0.1728197141 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 [Vitis vinifera] |
| 11 |
Hb_002249_070 |
0.174223347 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 12 |
Hb_004800_100 |
0.1753930238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005074_030 |
0.178129715 |
- |
- |
- |
| 14 |
Hb_005686_050 |
0.180065073 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 15 |
Hb_001504_330 |
0.180462887 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 16 |
Hb_158092_010 |
0.1808093397 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 17 |
Hb_001123_210 |
0.1822358477 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 18 |
Hb_007926_050 |
0.1838721028 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 19 |
Hb_009486_020 |
0.1842366255 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 20 |
Hb_000608_050 |
0.1852476513 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |