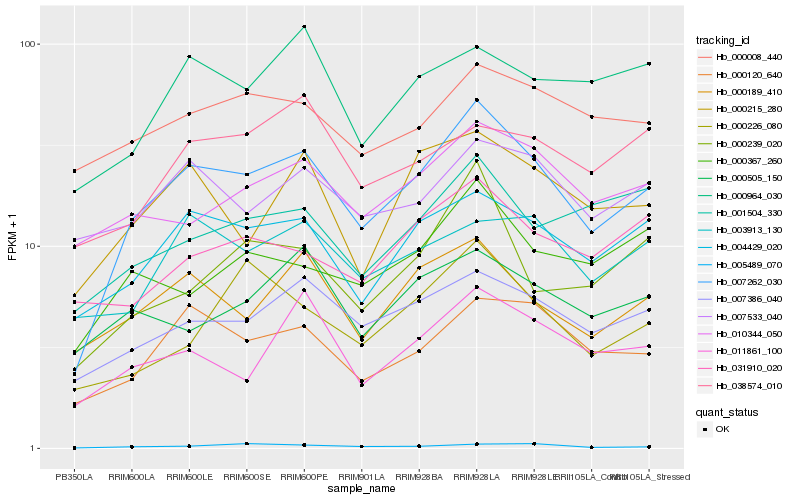
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007262_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_004429_020 |
0.1251842611 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 3 |
Hb_007386_040 |
0.1267548392 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000120_640 |
0.1319865488 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000189_410 |
0.1360527592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000215_280 |
0.142763333 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 7 |
Hb_011861_100 |
0.1446302351 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_010344_050 |
0.1474973918 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 9 |
Hb_000239_020 |
0.1506063474 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 10 |
Hb_000367_260 |
0.1512949388 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 11 |
Hb_007533_040 |
0.1537527006 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_003913_130 |
0.1555861047 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 13 |
Hb_000226_080 |
0.1557971027 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000008_440 |
0.1572092985 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 15 |
Hb_000964_030 |
0.1581693439 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 16 |
Hb_000505_150 |
0.1584436533 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 17 |
Hb_031910_020 |
0.1587594018 |
- |
- |
PREDICTED: exocyst complex component SEC15A [Jatropha curcas] |
| 18 |
Hb_001504_330 |
0.1587815025 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 19 |
Hb_005489_070 |
0.159285276 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 20 |
Hb_038574_010 |
0.1637883713 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |