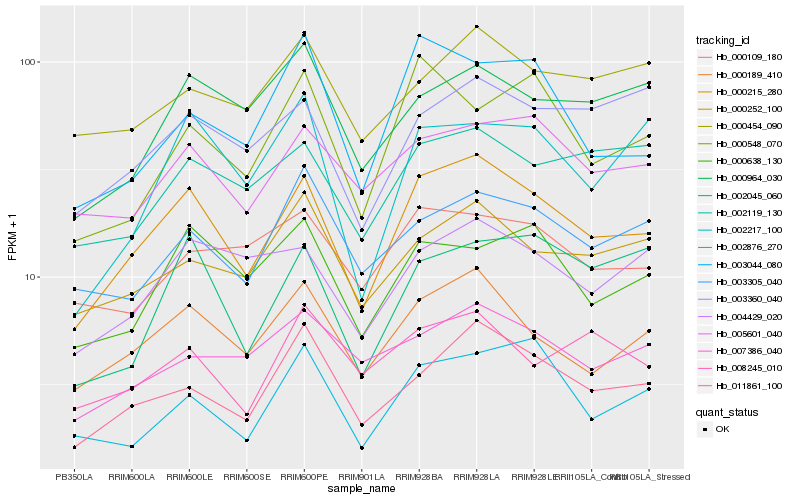
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_280 |
0.0 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_000189_410 |
0.0919448614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011861_100 |
0.1047531557 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_005601_040 |
0.119254247 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000638_130 |
0.1197149304 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 6 |
Hb_004429_020 |
0.1210237595 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 7 |
Hb_000252_100 |
0.1228576174 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 8 |
Hb_002119_130 |
0.1265671382 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 9 |
Hb_003305_040 |
0.1273112945 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 10 |
Hb_003044_080 |
0.127463075 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 11 |
Hb_003360_040 |
0.1290226517 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 12 |
Hb_000964_030 |
0.1305080374 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 13 |
Hb_002217_100 |
0.1312059363 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_002045_060 |
0.1316586195 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_007386_040 |
0.1318819444 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000109_180 |
0.1327296321 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 17 |
Hb_008245_010 |
0.1333854027 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 18 |
Hb_000454_090 |
0.1334263852 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 19 |
Hb_000548_070 |
0.133946543 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 20 |
Hb_002876_270 |
0.1355875913 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |