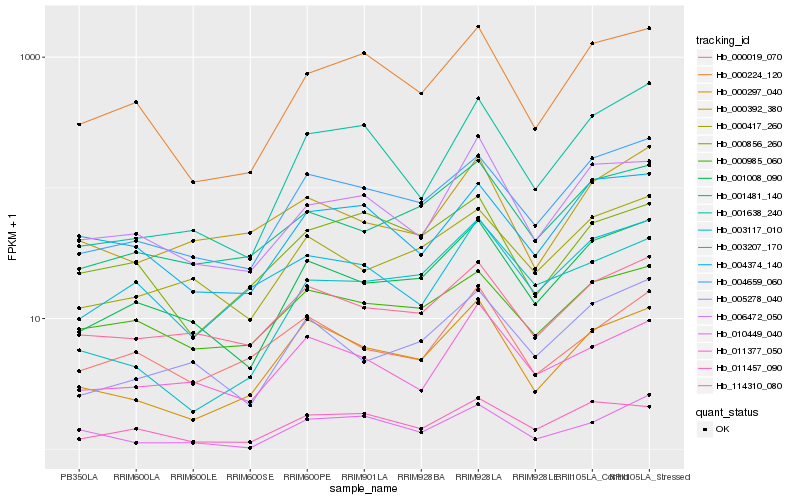
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000297_040 |
0.0 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 2 |
Hb_004659_060 |
0.1114400283 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 3 |
Hb_114310_080 |
0.1263065021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000224_120 |
0.12797262 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 5 |
Hb_001008_090 |
0.1279740218 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 6 |
Hb_000019_070 |
0.1312211412 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 7 |
Hb_011377_050 |
0.13405497 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000856_260 |
0.136664329 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 9 |
Hb_001481_140 |
0.1412334969 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 10 |
Hb_003207_170 |
0.1423245954 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006472_050 |
0.1425303735 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 12 |
Hb_011457_090 |
0.143292789 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000392_380 |
0.1438058225 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 14 |
Hb_000985_060 |
0.1479352592 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001638_240 |
0.1483663013 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 16 |
Hb_000417_260 |
0.1526298323 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_005278_040 |
0.1533595609 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 18 |
Hb_004374_140 |
0.1535751173 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 19 |
Hb_003117_010 |
0.1539272713 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 20 |
Hb_010449_040 |
0.1539342808 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |