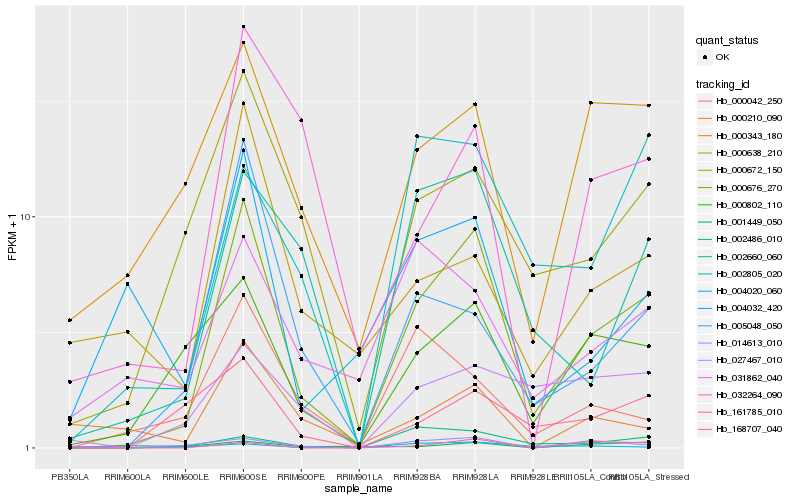
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_270 |
0.0 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 2 |
Hb_000802_110 |
0.2375720541 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 3 |
Hb_000672_150 |
0.2401716166 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 4 |
Hb_002660_060 |
0.2476805459 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 5 |
Hb_168707_040 |
0.2616729069 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000343_180 |
0.263687329 |
- |
- |
NDWp3 [Podospora anserina] |
| 7 |
Hb_005048_050 |
0.2669071403 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 8 |
Hb_002805_020 |
0.2744619662 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004032_420 |
0.2795947228 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 10 |
Hb_027467_010 |
0.2799001157 |
- |
- |
hypothetical protein PRUPE_ppa004106mg [Prunus persica] |
| 11 |
Hb_014613_010 |
0.2832214523 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 12 |
Hb_032264_090 |
0.2856057341 |
- |
- |
hypothetical protein JCGZ_02512 [Jatropha curcas] |
| 13 |
Hb_161785_010 |
0.2861503799 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 14 |
Hb_000042_250 |
0.293331388 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001449_050 |
0.2935861737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002486_010 |
0.2957653238 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |
| 17 |
Hb_000638_210 |
0.2958715705 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000210_090 |
0.2982188862 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 19 |
Hb_031862_040 |
0.2984097639 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 20 |
Hb_004020_060 |
0.2984306916 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |