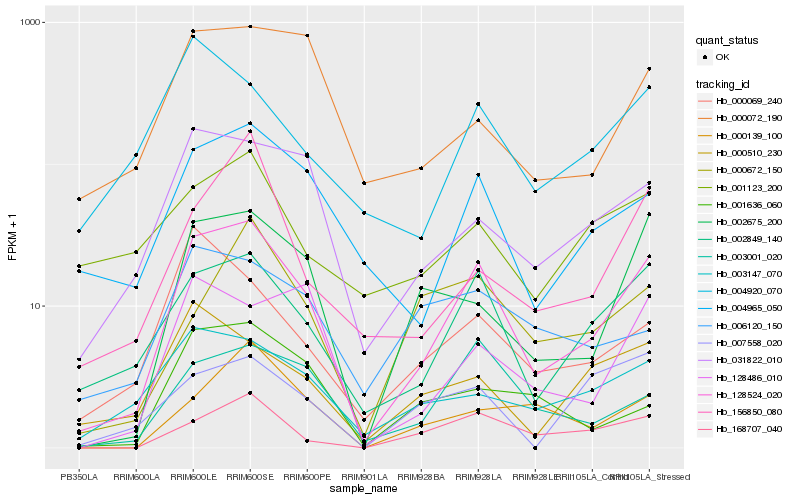
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128524_020 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002849_140 |
0.1589622098 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 3 |
Hb_004965_050 |
0.1959905156 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 4 |
Hb_002675_200 |
0.201934477 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 5 |
Hb_031822_010 |
0.2059898079 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 6 |
Hb_003001_020 |
0.2137109452 |
- |
- |
hypothetical protein JCGZ_17508 [Jatropha curcas] |
| 7 |
Hb_168707_040 |
0.2173952192 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_003147_070 |
0.2200068384 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 9 |
Hb_000510_230 |
0.2382504621 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 10 |
Hb_004920_070 |
0.239308882 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 11 |
Hb_128486_010 |
0.2412921065 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 12 |
Hb_000072_190 |
0.2416010586 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 13 |
Hb_156850_080 |
0.2426139085 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 14 |
Hb_001636_060 |
0.243366843 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 15 |
Hb_000139_100 |
0.2478332412 |
- |
- |
PREDICTED: sugar transport protein 13-like [Jatropha curcas] |
| 16 |
Hb_000069_240 |
0.2496488388 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 17 |
Hb_000672_150 |
0.2513829353 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 18 |
Hb_007558_020 |
0.2539843371 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 19 |
Hb_001123_200 |
0.2542735193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006120_150 |
0.2555228649 |
- |
- |
invertase [Hevea brasiliensis] |