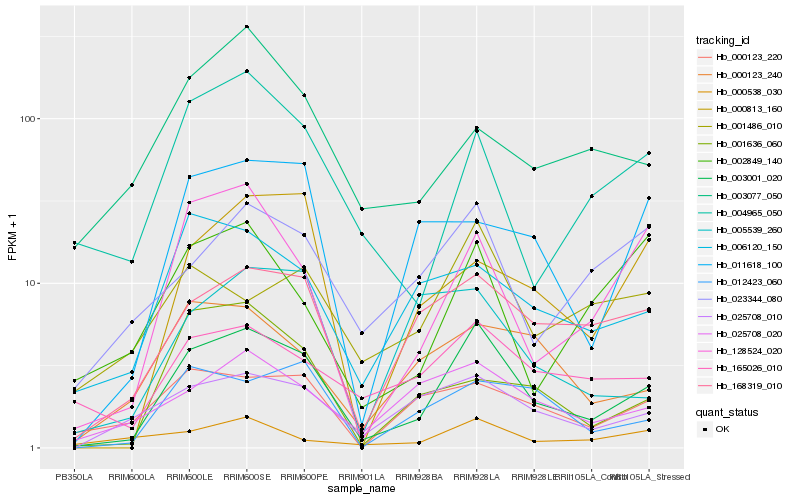
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003001_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_17508 [Jatropha curcas] |
| 2 |
Hb_168319_010 |
0.2023596811 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_128524_020 |
0.2137109452 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_025708_010 |
0.2143536971 |
- |
- |
hypothetical protein MTR_1g051060 [Medicago truncatula] |
| 5 |
Hb_025708_020 |
0.2228514304 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_012423_060 |
0.2247262759 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 7 |
Hb_165026_010 |
0.2261907141 |
transcription factor |
TF Family: NAC |
hypothetical protein JCGZ_14166 [Jatropha curcas] |
| 8 |
Hb_004965_050 |
0.2307343398 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 9 |
Hb_006120_150 |
0.23232321 |
- |
- |
invertase [Hevea brasiliensis] |
| 10 |
Hb_000123_240 |
0.2324125476 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_023344_080 |
0.2367200561 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000813_160 |
0.2416328639 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 13 |
Hb_011618_100 |
0.2427945827 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 14 |
Hb_001486_010 |
0.2438889564 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 15 |
Hb_000123_220 |
0.2444853806 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 16 |
Hb_000538_030 |
0.2499209102 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 17 |
Hb_001636_060 |
0.2523092351 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 18 |
Hb_005539_260 |
0.2529442256 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 19 |
Hb_002849_140 |
0.2550857403 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 20 |
Hb_003077_050 |
0.2618123536 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |