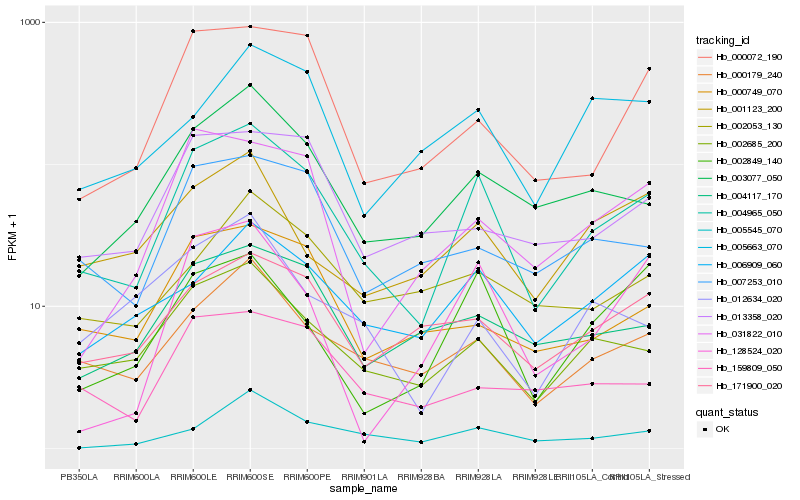
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004965_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 2 |
Hb_002685_200 |
0.1460757228 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 3 |
Hb_000179_240 |
0.1563194501 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003077_050 |
0.1585825851 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 5 |
Hb_000072_190 |
0.1769665109 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 6 |
Hb_004117_170 |
0.1784994211 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 7 |
Hb_171900_020 |
0.1838418444 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 8 |
Hb_000749_070 |
0.1840268673 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 9 |
Hb_007253_010 |
0.1844294444 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 10 |
Hb_006909_060 |
0.1854906709 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 11 |
Hb_002849_140 |
0.1893949789 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 12 |
Hb_159809_050 |
0.1914394048 |
- |
- |
AT14A, putative [Ricinus communis] |
| 13 |
Hb_031822_010 |
0.191870231 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 14 |
Hb_013358_020 |
0.1956951738 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 15 |
Hb_128524_020 |
0.1959905156 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005545_070 |
0.2021415257 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 17 |
Hb_001123_200 |
0.2030780736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002053_130 |
0.2049064836 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 19 |
Hb_012634_020 |
0.2058387598 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005663_070 |
0.2082145512 |
- |
- |
dehydrin protein [Manihot esculenta] |