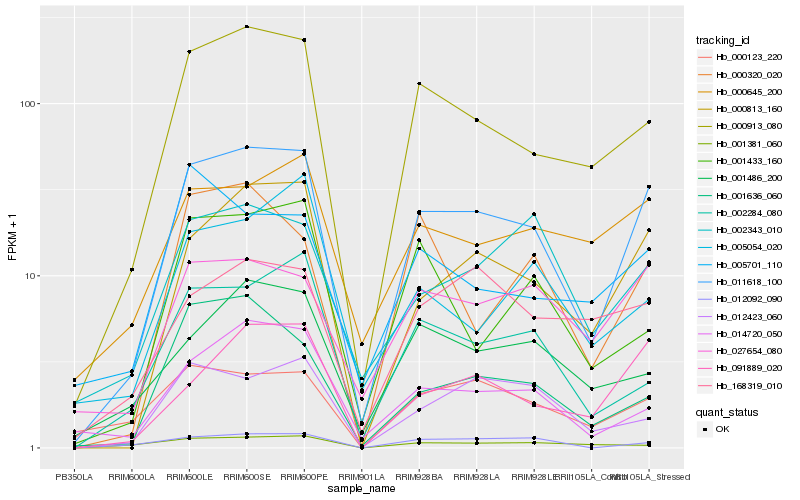
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_100 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 2 |
Hb_000913_080 |
0.128718517 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 3 |
Hb_000813_160 |
0.1352721339 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 4 |
Hb_091889_020 |
0.166650869 |
- |
- |
- |
| 5 |
Hb_027654_080 |
0.1732262425 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_014720_050 |
0.1766433427 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 7 |
Hb_002284_080 |
0.1810326464 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000645_200 |
0.1813876078 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 9 |
Hb_012092_090 |
0.1814249736 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012423_060 |
0.1864327701 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 11 |
Hb_001486_200 |
0.1869272148 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 12 |
Hb_000123_220 |
0.1879278925 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 13 |
Hb_002343_010 |
0.1889780329 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_001636_060 |
0.1984056321 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 15 |
Hb_168319_010 |
0.1990614001 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000320_020 |
0.2003834984 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 17 |
Hb_001433_160 |
0.2048026303 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 18 |
Hb_001381_060 |
0.2055857115 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 19 |
Hb_005701_110 |
0.205699195 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 20 |
Hb_005054_020 |
0.2094192335 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |