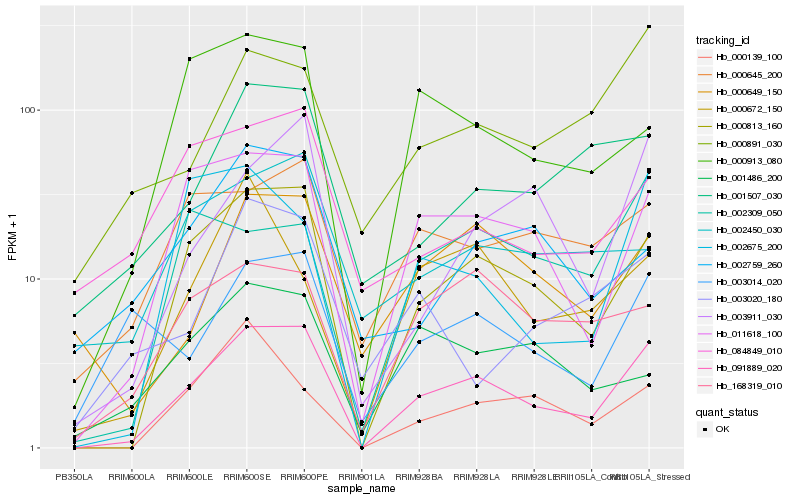
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091889_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000813_160 |
0.1040125314 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 3 |
Hb_011618_100 |
0.166650869 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 4 |
Hb_003911_030 |
0.2102404813 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000913_080 |
0.215522509 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 6 |
Hb_000649_150 |
0.2159335414 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 7 |
Hb_168319_010 |
0.2181237644 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000645_200 |
0.2205266333 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 9 |
Hb_001507_030 |
0.2219488343 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 10 |
Hb_000891_030 |
0.2246866274 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 11 |
Hb_003014_020 |
0.22510219 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 12 |
Hb_002675_200 |
0.237041561 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 13 |
Hb_000139_100 |
0.2375380449 |
- |
- |
PREDICTED: sugar transport protein 13-like [Jatropha curcas] |
| 14 |
Hb_002309_050 |
0.2383387299 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 15 |
Hb_002450_030 |
0.2405697641 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 16 |
Hb_003020_180 |
0.2421647256 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 17 |
Hb_084849_010 |
0.242623535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001486_200 |
0.243230822 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 19 |
Hb_000672_150 |
0.2435930843 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 20 |
Hb_002759_260 |
0.2441314609 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |