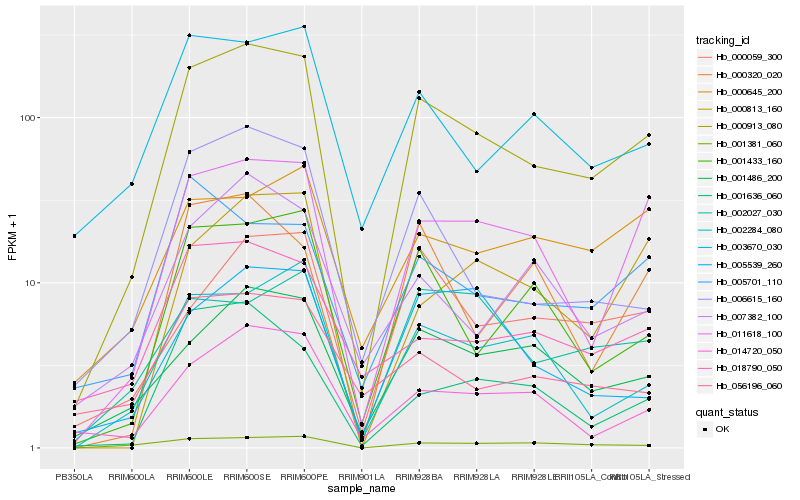
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000913_080 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 2 |
Hb_011618_100 |
0.128718517 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 3 |
Hb_001486_200 |
0.1476574308 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 4 |
Hb_001433_160 |
0.1589189196 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 5 |
Hb_001381_060 |
0.1611519633 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 6 |
Hb_014720_050 |
0.1618335397 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 7 |
Hb_002284_080 |
0.1646661444 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_003670_030 |
0.1671188078 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 9 |
Hb_000059_300 |
0.171571148 |
- |
- |
caspase, putative [Ricinus communis] |
| 10 |
Hb_006615_160 |
0.171755315 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 11 |
Hb_018790_050 |
0.1779586156 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 12 |
Hb_001636_060 |
0.178413198 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 13 |
Hb_000813_160 |
0.1821365645 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 14 |
Hb_007382_100 |
0.1829050466 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_005701_110 |
0.1838520549 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 16 |
Hb_002027_030 |
0.1857339583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000645_200 |
0.1869800394 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 18 |
Hb_000320_020 |
0.1876670528 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 19 |
Hb_056196_060 |
0.1880673675 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_005539_260 |
0.1889729585 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |