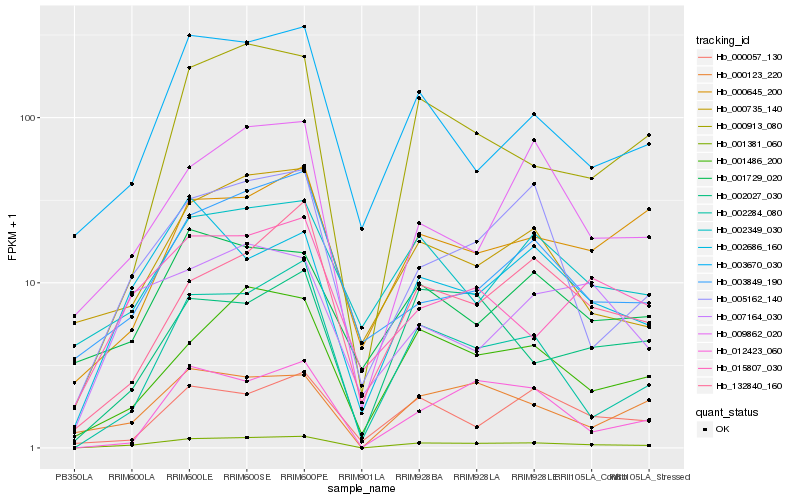
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001381_060 |
0.0 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 2 |
Hb_001486_200 |
0.1542128674 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 3 |
Hb_002284_080 |
0.159688602 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000913_080 |
0.1611519633 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 5 |
Hb_002686_160 |
0.1712930245 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 6 |
Hb_000123_220 |
0.1725564329 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 7 |
Hb_003670_030 |
0.1725618884 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 8 |
Hb_005162_140 |
0.1752775302 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 9 |
Hb_015807_030 |
0.1762701114 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 10 |
Hb_000735_140 |
0.1780696896 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 11 |
Hb_012423_060 |
0.1800772255 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 12 |
Hb_132840_160 |
0.1823625236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009862_020 |
0.1833539298 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 14 |
Hb_002027_030 |
0.1839003474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002349_030 |
0.1845053832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000645_200 |
0.1848758864 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 17 |
Hb_003849_190 |
0.1863103221 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 18 |
Hb_000057_130 |
0.1869542297 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 19 |
Hb_007164_030 |
0.1876248572 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 20 |
Hb_001729_020 |
0.1879274654 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |