| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
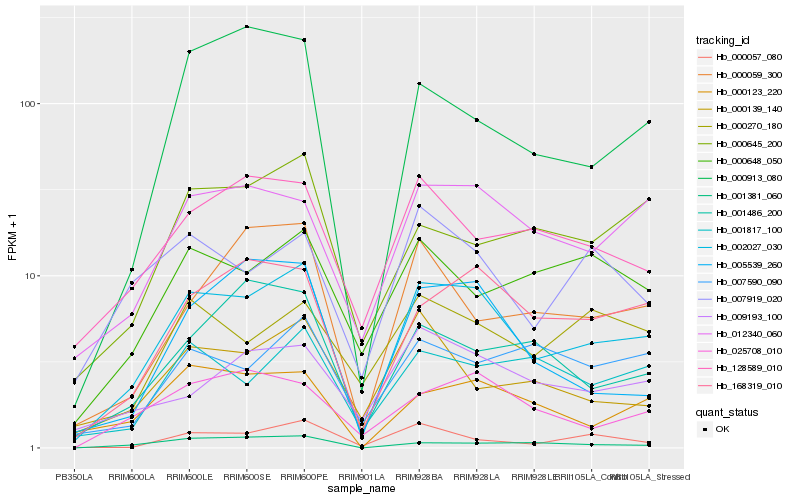
| 1 |
Hb_002027_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_168319_010 |
0.1484429899 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_012340_060 |
0.1604197003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000123_220 |
0.1705711485 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 5 |
Hb_025708_010 |
0.1743209476 |
- |
- |
hypothetical protein MTR_1g051060 [Medicago truncatula] |
| 6 |
Hb_005539_260 |
0.1784242962 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 7 |
Hb_001817_100 |
0.1801567655 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 8 |
Hb_128589_010 |
0.1815071382 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_001381_060 |
0.1839003474 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 10 |
Hb_000913_080 |
0.1857339583 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 11 |
Hb_000645_200 |
0.1886800134 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 12 |
Hb_000057_080 |
0.1888045774 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 13 |
Hb_009193_100 |
0.188817276 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 14 |
Hb_000270_180 |
0.1895325164 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 15 |
Hb_007919_020 |
0.1899900642 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 16 |
Hb_007590_090 |
0.1927189869 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 17 |
Hb_000059_300 |
0.1935811803 |
- |
- |
caspase, putative [Ricinus communis] |
| 18 |
Hb_000648_050 |
0.1937146999 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001486_200 |
0.1947579622 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 20 |
Hb_000139_140 |
0.2007286351 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |