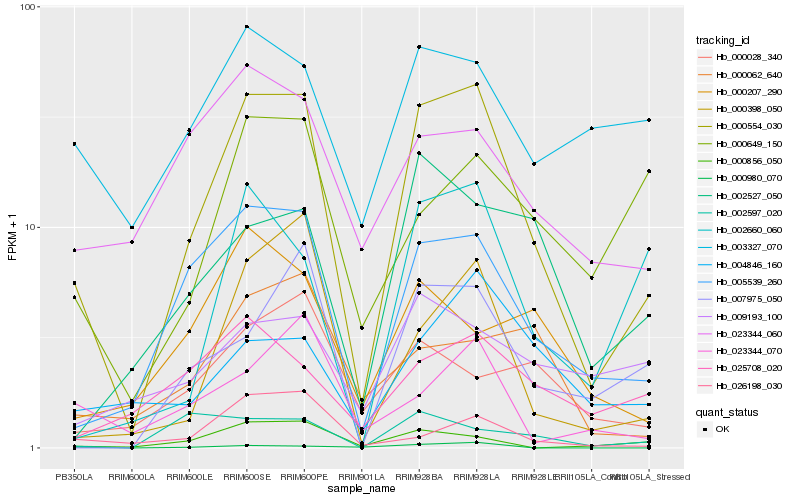
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000554_030 |
0.0 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005539_260 |
0.1790756075 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 3 |
Hb_000398_050 |
0.2272657652 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 4 |
Hb_007975_050 |
0.2379150128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 5 |
Hb_000856_050 |
0.24688574 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 6 |
Hb_002660_060 |
0.2494367062 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 7 |
Hb_002527_050 |
0.2571629123 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 8 |
Hb_026198_030 |
0.2577749452 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 9 |
Hb_004846_160 |
0.2596653508 |
- |
- |
- |
| 10 |
Hb_023344_060 |
0.2624118144 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 11 |
Hb_023344_070 |
0.2658077307 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 12 |
Hb_025708_020 |
0.27034627 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000062_640 |
0.2722088753 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000028_340 |
0.2732349368 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 15 |
Hb_003327_070 |
0.2735152596 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 16 |
Hb_000649_150 |
0.2740492642 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 17 |
Hb_000207_290 |
0.2753042999 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 18 |
Hb_002597_020 |
0.2776666101 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_009193_100 |
0.280395765 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 20 |
Hb_000980_070 |
0.2819153966 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |