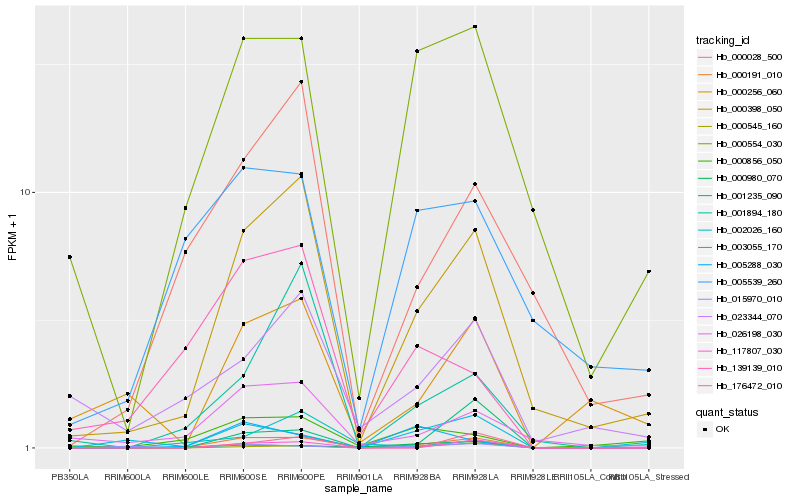
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_010 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_000398_050 |
0.1995755277 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 3 |
Hb_000545_160 |
0.2683417027 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 4 |
Hb_026198_030 |
0.2735264669 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 5 |
Hb_117807_030 |
0.2745634119 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 6 |
Hb_000554_030 |
0.2847875093 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000028_500 |
0.3006752606 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001235_090 |
0.3020233507 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 9 |
Hb_023344_070 |
0.3113425474 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_139139_010 |
0.3157935379 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000856_050 |
0.3205090788 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 12 |
Hb_015970_010 |
0.3223823239 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 13 |
Hb_000980_070 |
0.3249741247 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 14 |
Hb_001894_180 |
0.3258271351 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 15 |
Hb_000256_060 |
0.3267231061 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 16 |
Hb_176472_010 |
0.3287638144 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 17 |
Hb_005539_260 |
0.3315601226 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 18 |
Hb_003055_170 |
0.3330724491 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005288_030 |
0.3377996606 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 20 |
Hb_002026_160 |
0.3443569304 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |