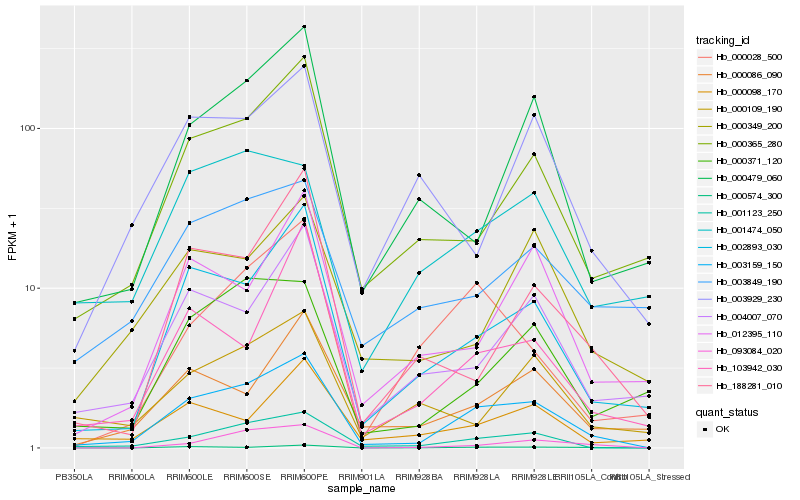
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003159_150 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 2 |
Hb_001123_250 |
0.14448225 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 3 |
Hb_002893_030 |
0.164118415 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 4 |
Hb_000365_280 |
0.1905488578 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 5 |
Hb_004007_070 |
0.2012039589 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 6 |
Hb_000371_120 |
0.2055402073 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000086_090 |
0.2095669117 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_093084_020 |
0.2101368283 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Populus euphratica] |
| 9 |
Hb_000479_060 |
0.2104581435 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 10 |
Hb_000574_300 |
0.2108967486 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 11 |
Hb_000028_500 |
0.216525646 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000349_200 |
0.2181070204 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 13 |
Hb_012395_110 |
0.2205340825 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 14 |
Hb_103942_030 |
0.2219783404 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000098_170 |
0.2224548827 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 16 |
Hb_003849_190 |
0.2231677388 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 17 |
Hb_000109_190 |
0.2271824665 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 18 |
Hb_188281_010 |
0.2272101017 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 19 |
Hb_003929_230 |
0.2325851099 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 20 |
Hb_001474_050 |
0.2329778499 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |